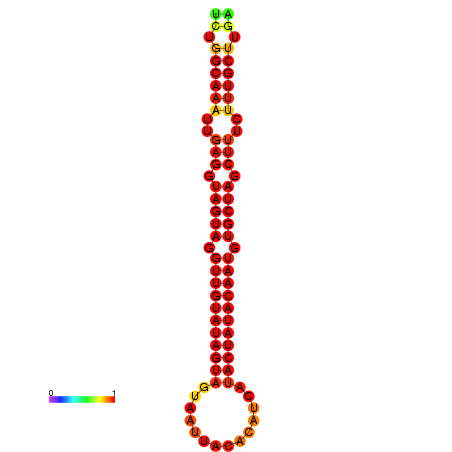
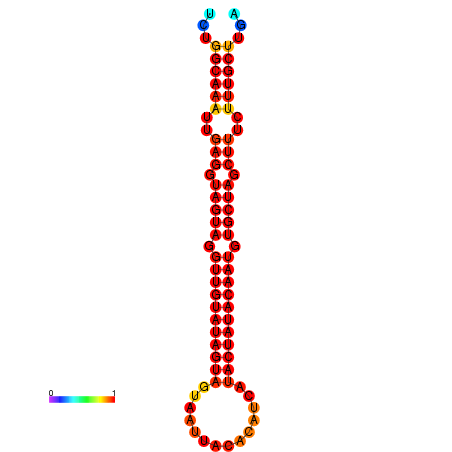
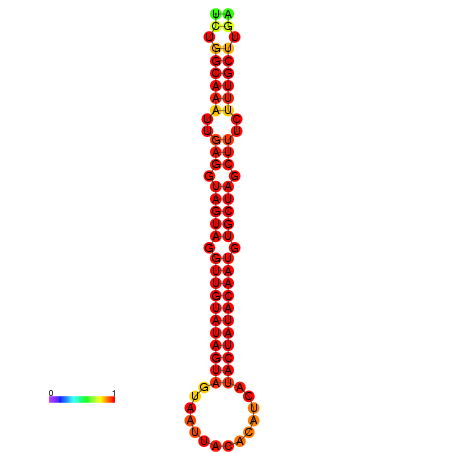
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:18472019-18472126 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droSim1 | chr2L:18160930-18161037 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droSec1 | super_7:2097967-2098074 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droYak2 | chr2R:4988819-4988926 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droEre2 | scaffold_4845:5799401-5799508 - | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droAna3 | scaffold_12916:2379166-2379273 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGCAACTA-AGAATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACAATAAGCCGCATTT |
| dp4 | chr4_group3:3635640-3635746 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTT--CAGATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCTCATTT |
| droPer1 | super_1:5116219-5116325 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTT--CAGATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCTCATTT |
| droWil1 | scaffold_180708:11623134-11623240 + | AAAGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTATTCAATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCACA--- |
| droVir3 | scaffold_12963:18451337-18451441 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTATTTA-TAGAGCATACTATACAGCGTGCTAGCTTTCTTTGCTTGACAATAAGCCGCA--- |
| droMoj3 | scaffold_6500:6184568-6184672 - | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTTA-TAGACCATACTATACAGCGTGCTAGCTTTCTTTGCTTGACTATAAGCCGCA--- |
| droGri2 | scaffold_3792:132-239 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTATCTA-TCTAGCATACTATACAGCTTGCTAGCTTTCTTTGCTTGACTATAAGCAGCATAG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:18472019-18472126 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droSim1 | chr2L:18160930-18161037 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droSec1 | super_7:2097967-2098074 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droYak2 | chr2R:4988819-4988926 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droEre2 | scaffold_4845:5799401-5799508 - | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTA-CACATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTACAAGCCGCATTT |
| droAna3 | scaffold_12916:2379166-2379273 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGCAACTA-AGAATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACAATAAGCCGCATTT |
| dp4 | chr4_group3:3635640-3635746 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTT--CAGATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCTCATTT |
| droPer1 | super_1:5116219-5116325 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTT--CAGATCGTACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCTCATTT |
| droWil1 | scaffold_180708:11623134-11623240 + | AAAGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTAATTATTCAATCATACTATACAATGTGCTAGCTTTCTTTGCTTGACTATAAGCCACA--- |
| droVir3 | scaffold_12963:18451337-18451441 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTATTTA-TAGAGCATACTATACAGCGTGCTAGCTTTCTTTGCTTGACAATAAGCCGCA--- |
| droMoj3 | scaffold_6500:6184568-6184672 - | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTACTTA-TAGACCATACTATACAGCGTGCTAGCTTTCTTTGCTTGACTATAAGCCGCA--- |
| droGri2 | scaffold_3792:132-239 + | A-AGGACCTTTTTCTCTCTGGCAAATTGAGGTAGTAGGTTGTATAGTAGTATCTA-TCTAGCATACTATACAGCTTGCTAGCTTTCTTTGCTTGACTATAAGCAGCATAG |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -30.2 | -29.5 |
|---|---|
 |
 |
| -30.2 | -29.5 |
|---|---|
 |
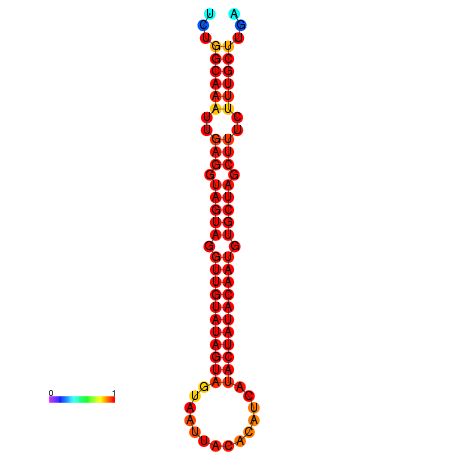 |
| -30.2 | -29.5 |
|---|---|
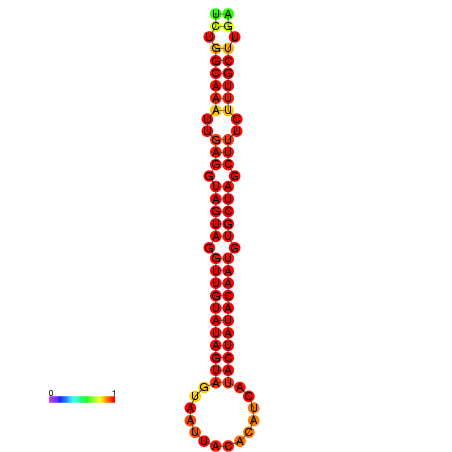 |
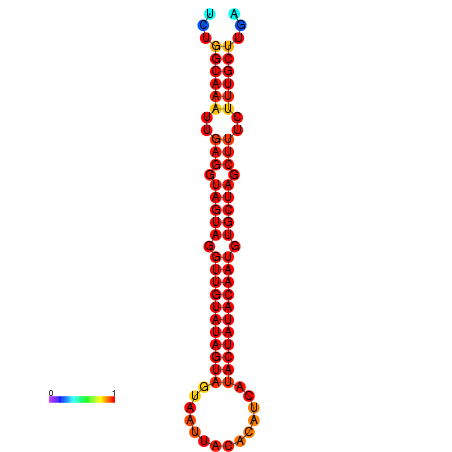 |
| -30.2 | -29.5 |
|---|---|
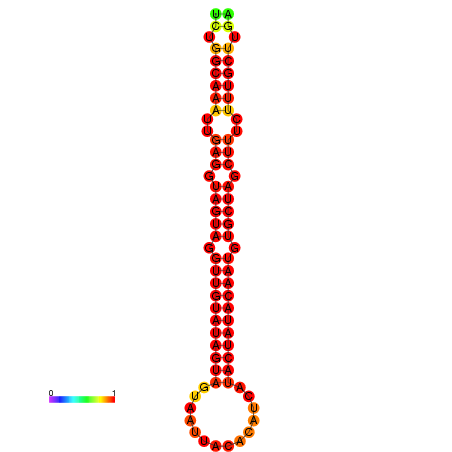 |
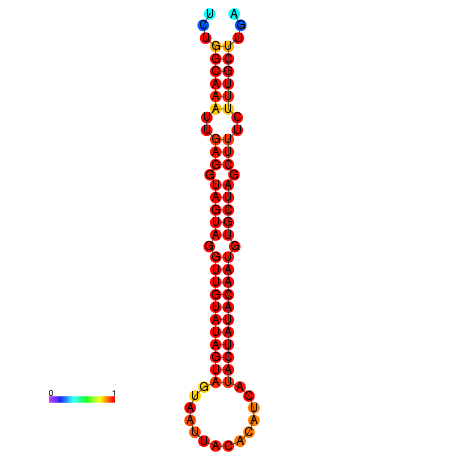 |
| -30.2 | -29.5 |
|---|---|
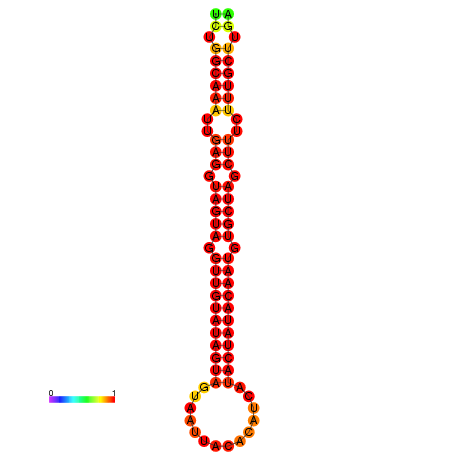 |
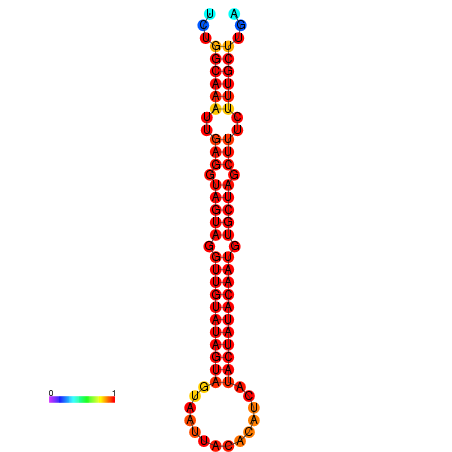 |
| -30.1 | -29.4 | -29.2 |
|---|---|---|
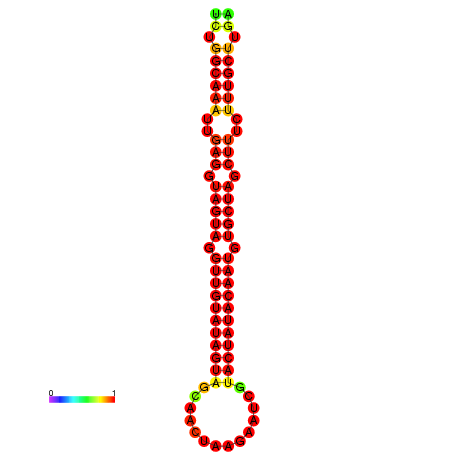 |
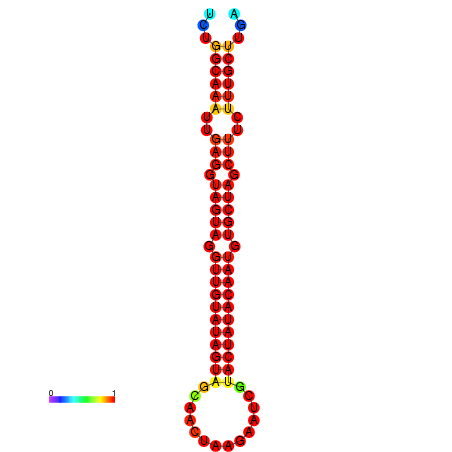 |
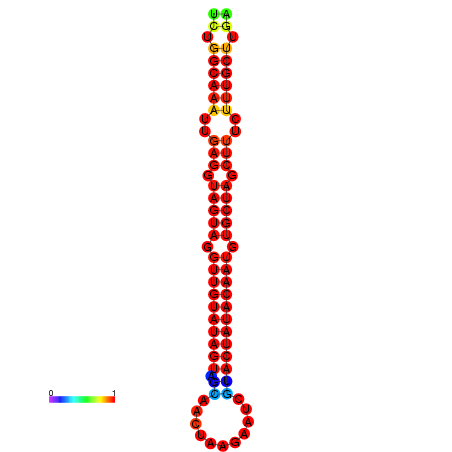 |
| -30.2 | -29.5 |
|---|---|
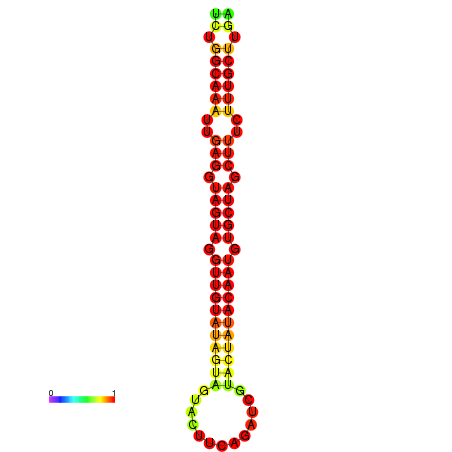 |
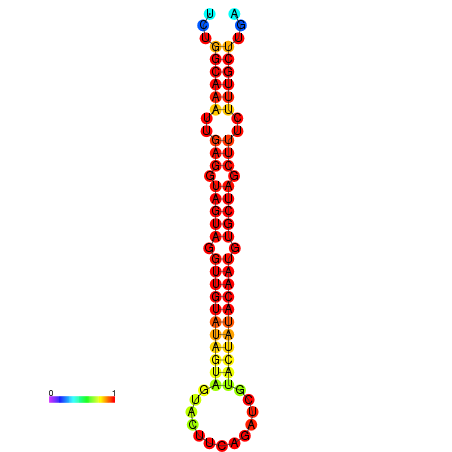 |
| -30.2 | -29.5 |
|---|---|
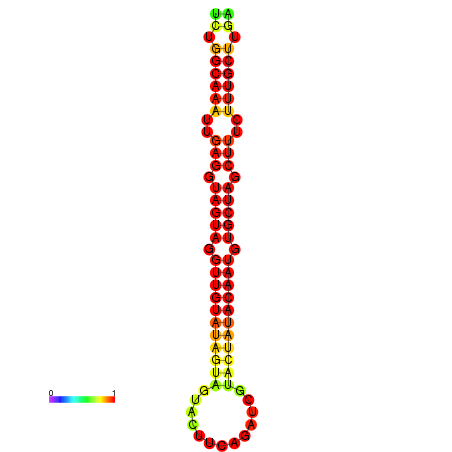 |
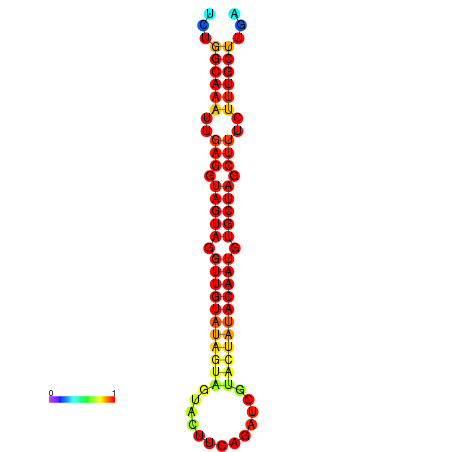 |
| -30.2 | -29.7 | -29.5 |
|---|---|---|
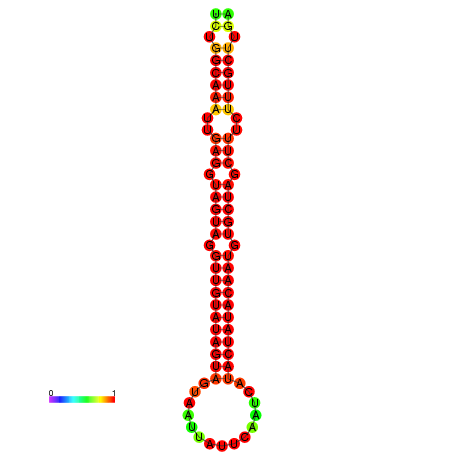 |
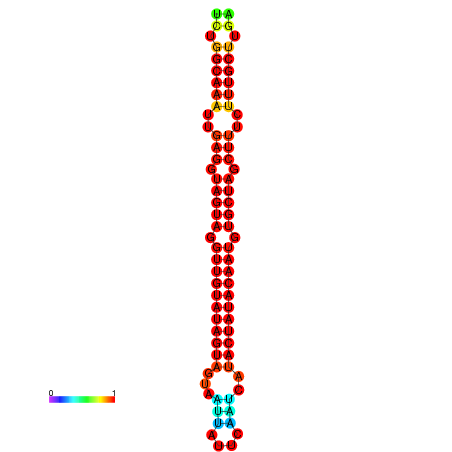 |
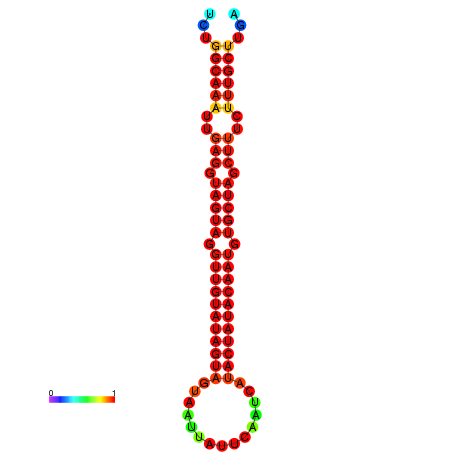 |
| -32.3 | -31.9 | -31.7 | -31.6 |
|---|---|---|---|
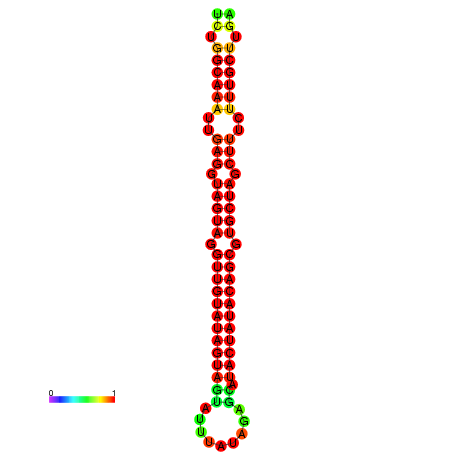 |
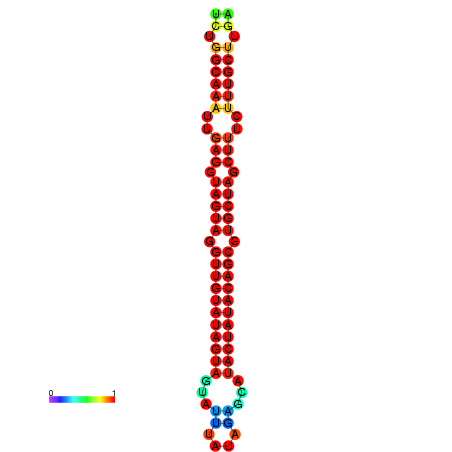 |
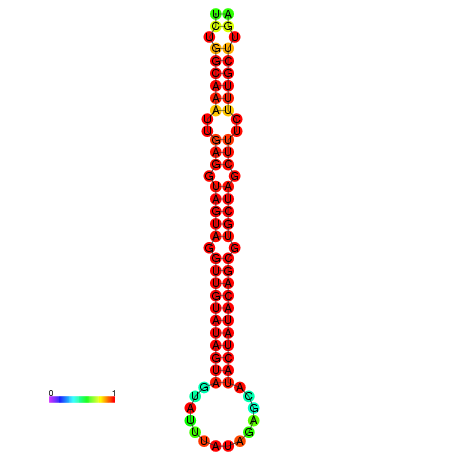 |
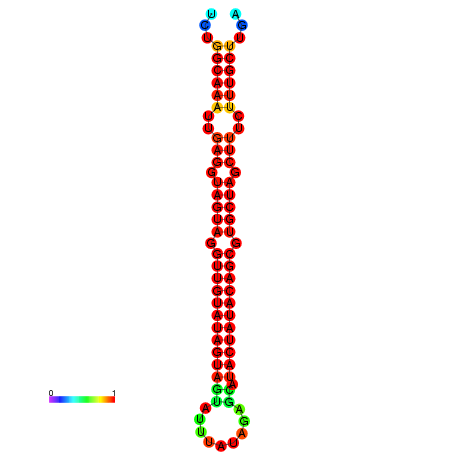 |
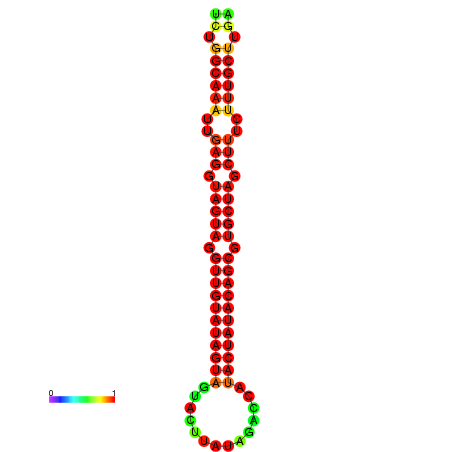
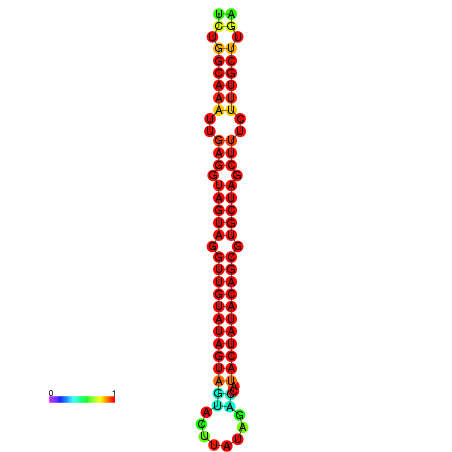
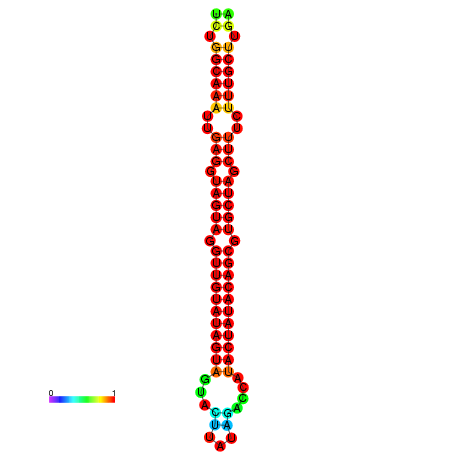
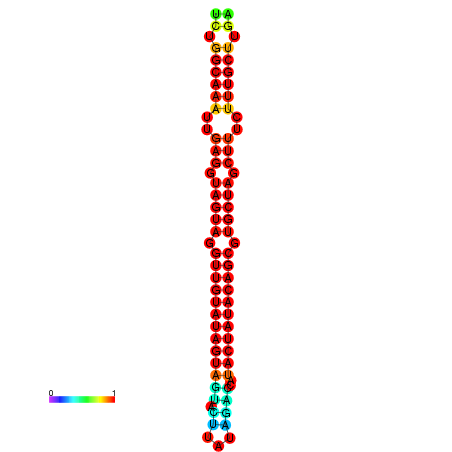
| -31.7 | -31.3 | -31.2 | -31.1 | -31.0 |
|---|---|---|---|---|
 |
 |
 |
 |
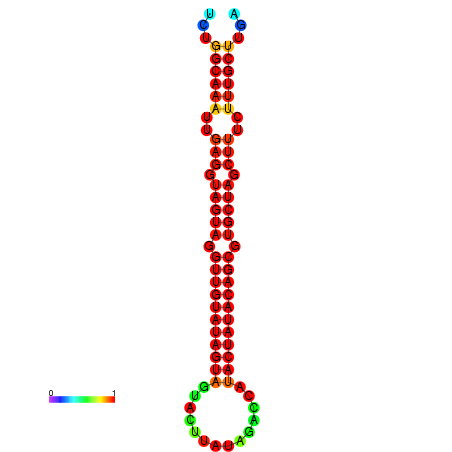 |
| -33.6 | -33.0 | -32.9 | -32.6 |
|---|---|---|---|
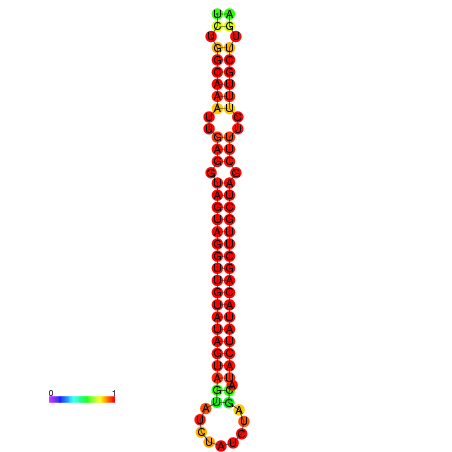 |
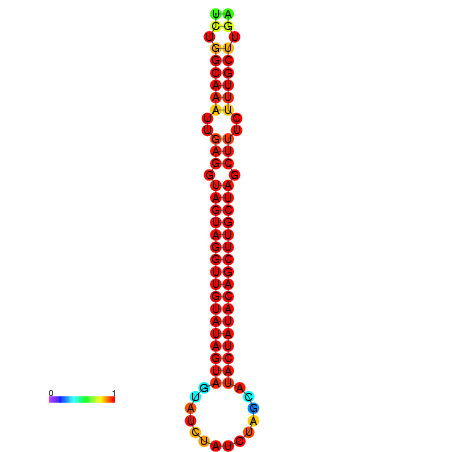 |
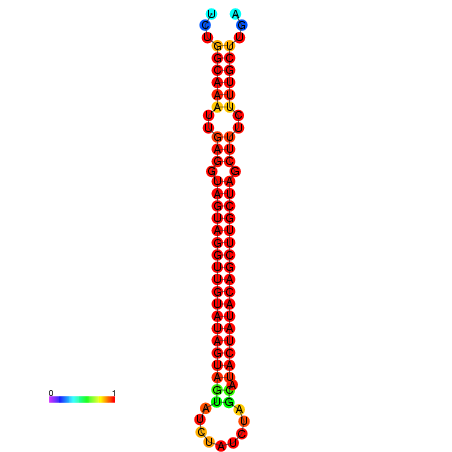 |
 |
Generated: 01/05/2013 at 05:04 PM