

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:642193-642303 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droSim1 | chr3L:290535-290645 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droSec1 | super_2:662642-662752 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droYak2 | chr3L:629639-629749 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droEre2 | scaffold_4784:648329-648439 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droAna3 | scaffold_13337:6520579-6520690 - | TTTGT---------------------------AAACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| dp4 | chrXR_group8:4603895-4604015 - | TTTGGGAC-------T-----ACTCCACCAAT---A---TATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droPer1 | super_29:844606-844726 - | TTTGGGAC-------T-----ACTCCACCAAT---A---TATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droWil1 | scaffold_180698:10172097-10172254 + | TTTTTTTCATACATTTT-------------------TTTGATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACAGAAAAAAAAAGAAGAAAAAAAAATAACGATCATCATCTTGCC |
| droVir3 | scaffold_13049:5805300-5805416 + | TTTGTGTG-------T-----CGTCT------A-TCTC---TGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTAT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droMoj3 | scaffold_6680:4143306-4143408 + | CT--------------------------------------ATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTTTTTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATAACCACA--------------------------------------TTCC |
| droGri2 | scaffold_15110:13265206-13265333 - | TGTGCGTC-------TCTCTGTCTCCATCACT---A-CCTATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCAGACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:642193-642303 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droSim1 | chr3L:290535-290645 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droSec1 | super_2:662642-662752 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droYak2 | chr3L:629639-629749 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droEre2 | scaffold_4784:648329-648439 + | TTTGT---------------------------A-ACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droAna3 | scaffold_13337:6520579-6520690 - | TTTGT---------------------------AAACTCCAATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| dp4 | chrXR_group8:4603895-4604015 - | TTTGGGAC-------T-----ACTCCACCAAT---A---TATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droPer1 | super_29:844606-844726 - | TTTGGGAC-------T-----ACTCCACCAAT---A---TATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droWil1 | scaffold_180698:10172097-10172254 + | TTTTTTTCATACATTTT-------------------TTTGATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACAGAAAAAAAAAGAAGAAAAAAAAATAACGATCATCATCTTGCC |
| droVir3 | scaffold_13049:5805300-5805416 + | TTTGTGTG-------T-----CGTCT------A-TCTC---TGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTAT-TTCATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| droMoj3 | scaffold_6680:4143306-4143408 + | CT--------------------------------------ATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTTTTTTATACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATAACCACA--------------------------------------TTCC |
| droGri2 | scaffold_15110:13265206-13265333 - | TGTGCGTC-------TCTCTGTCTCCATCACT---A-CCTATGATTTGACTACGAAACCGGTTTTCGATTTGGTTTGACTGTTT-TTCAGACAAGTGAGATCATTTTGAAAGCTGATTTTGTCAATGAATA-CCACA--------------------------------------TTCC |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
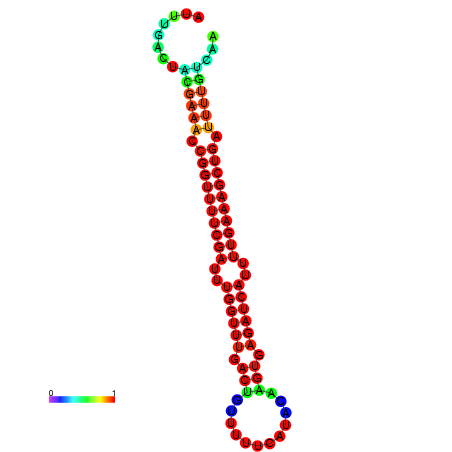 |
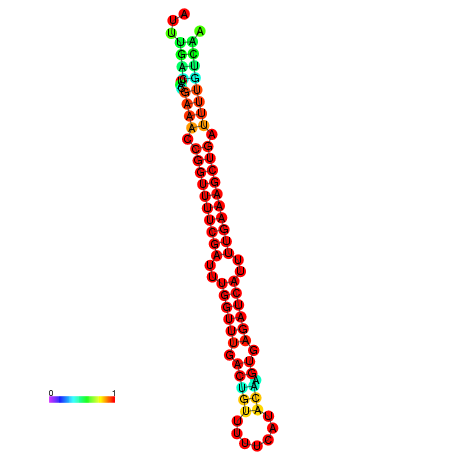 |
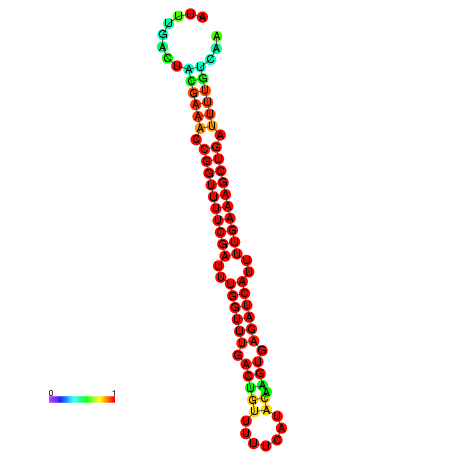
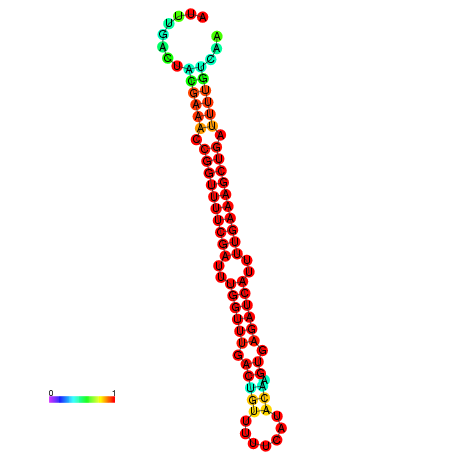
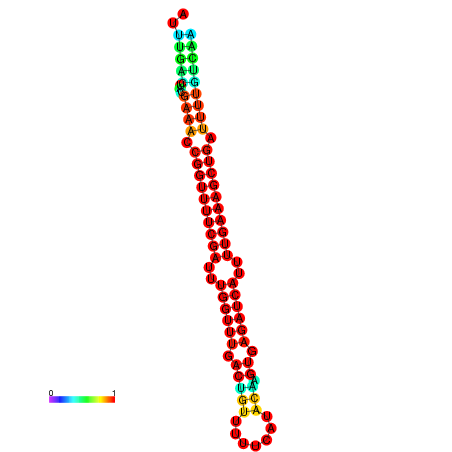
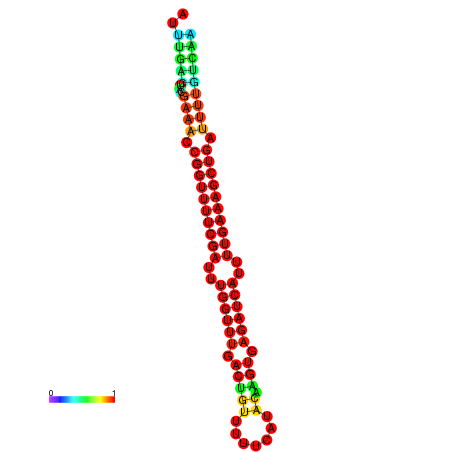
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
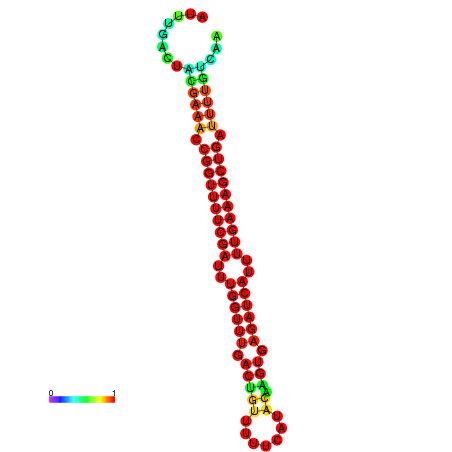 |
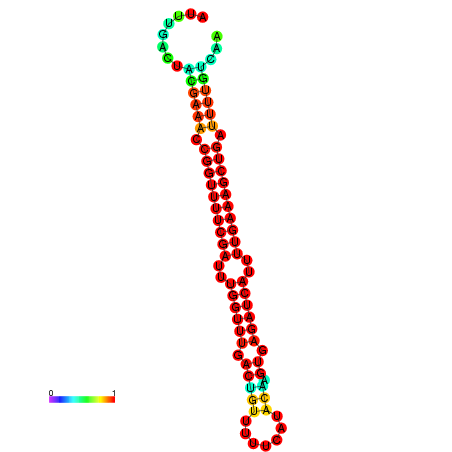 |
 |
 |
 |
 |
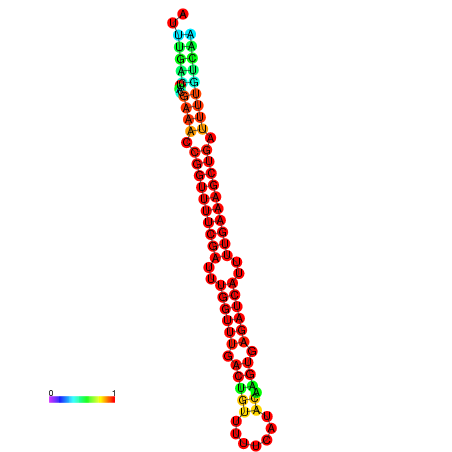
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
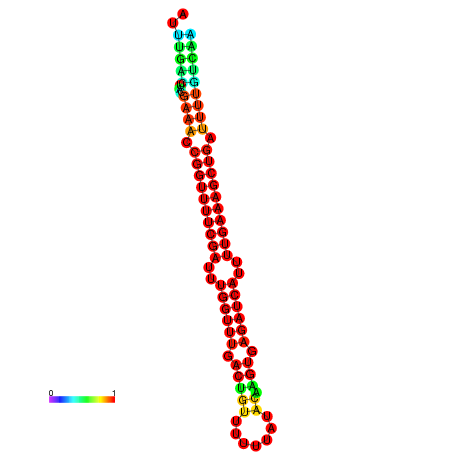
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
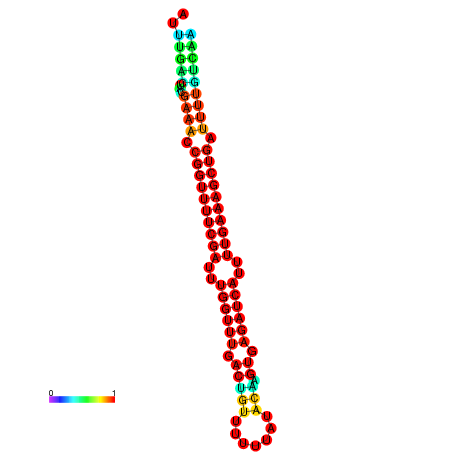
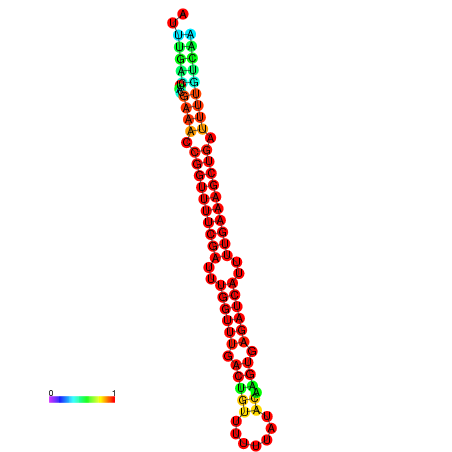
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -26.0 | -26.0 | -25.7 | -25.7 | -25.4 | -25.3 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
| -27.6 | -27.6 | -27.3 | -27.3 | -26.9 | -26.9 |
|---|---|---|---|---|---|
 |
 |
 |
 |
 |
 |
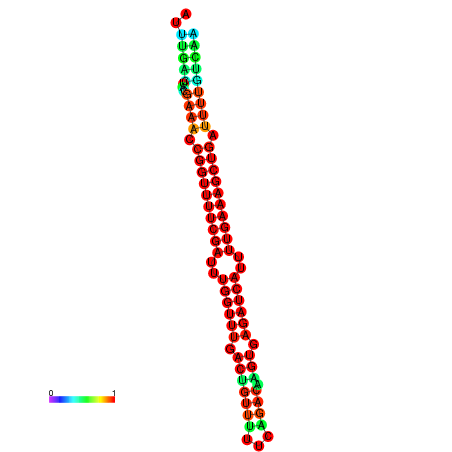
| -26.5 | -26.5 | -26.2 | -26.2 | -25.8 | -25.8 |
|---|---|---|---|---|---|
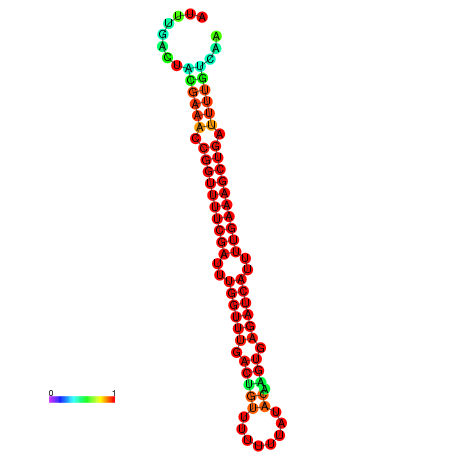 |
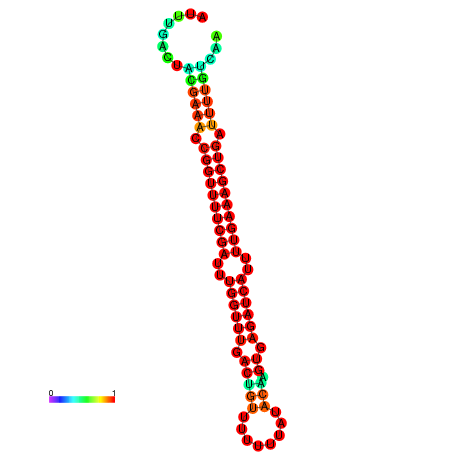 |
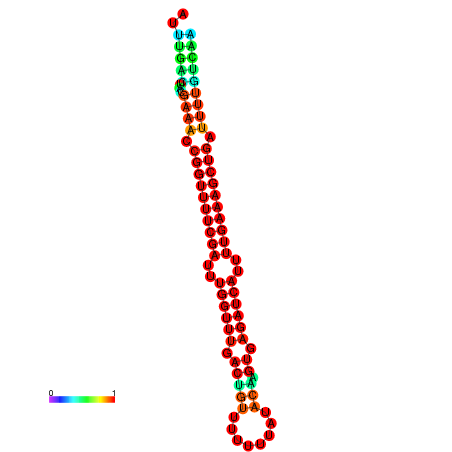 |
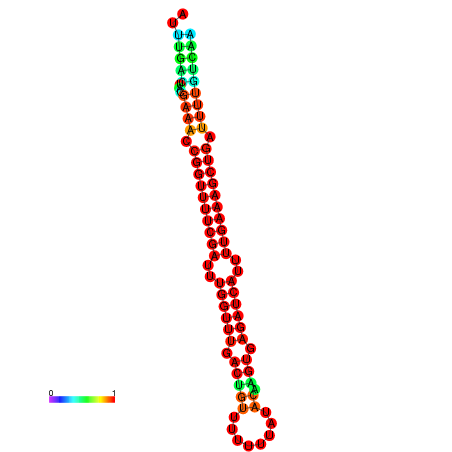 |
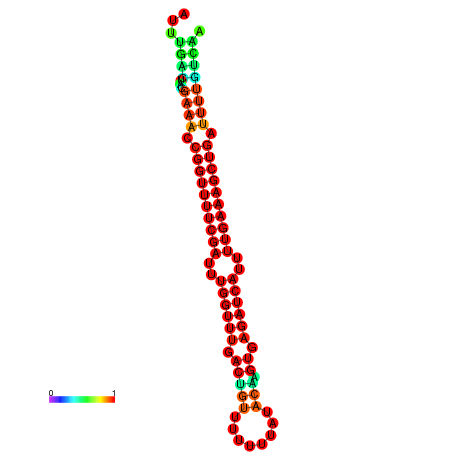 |
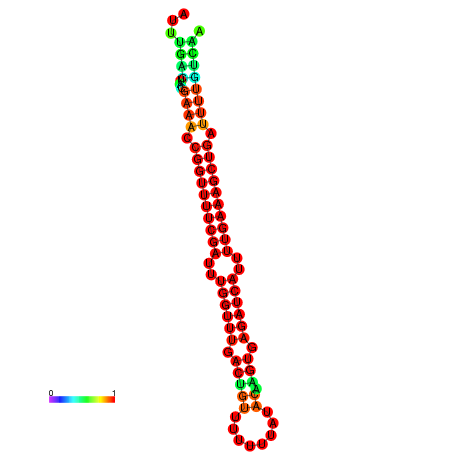 |
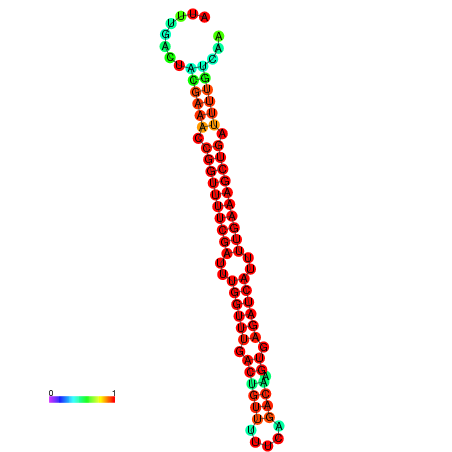
| -27.1 | -27.1 | -26.9 | -26.9 | -26.8 | -26.8 |
|---|---|---|---|---|---|
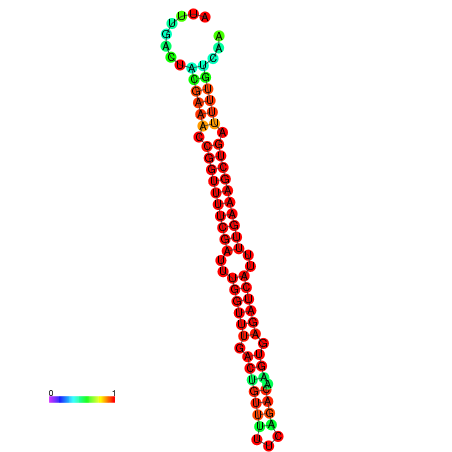 |
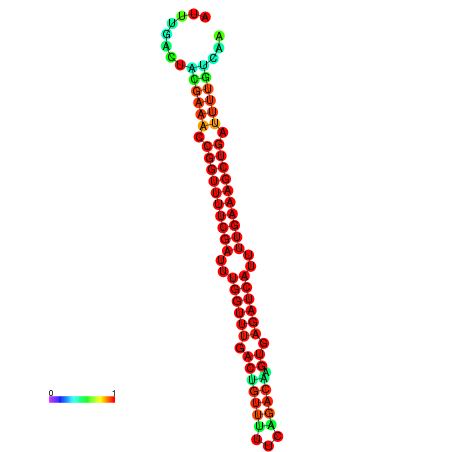 |
 |
 |
 |
 |
Generated: 01/05/2013 at 05:04 PM