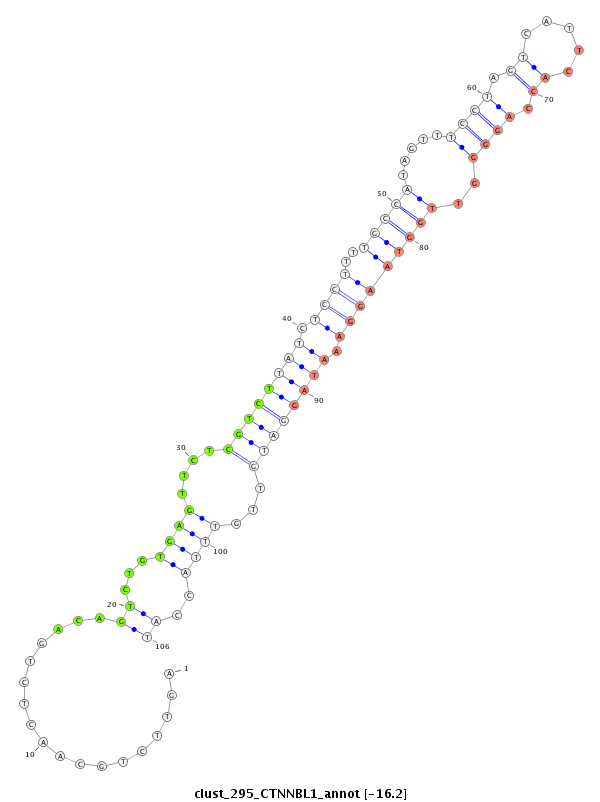
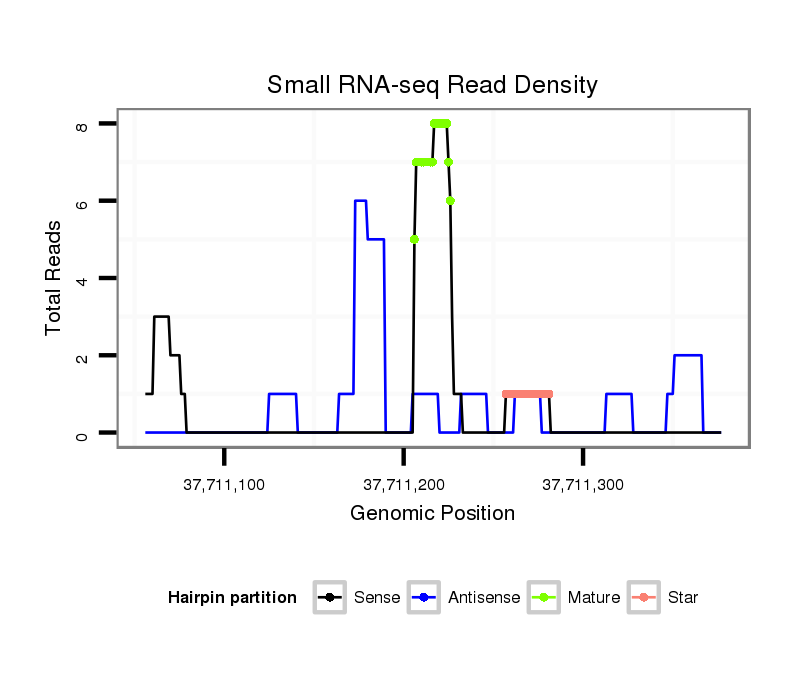
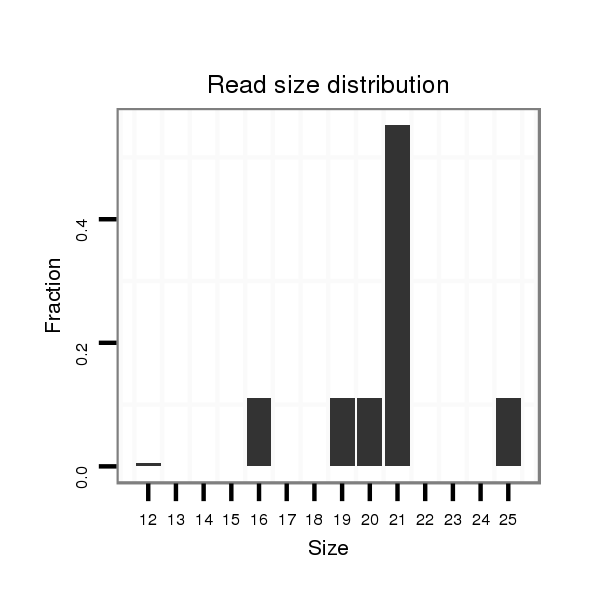
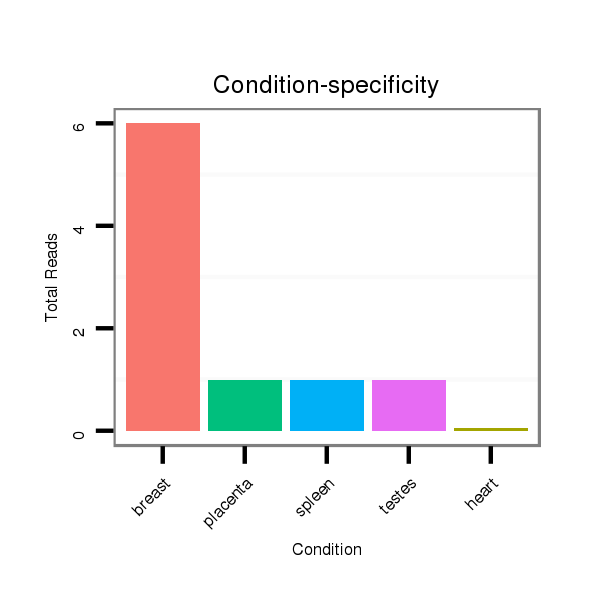
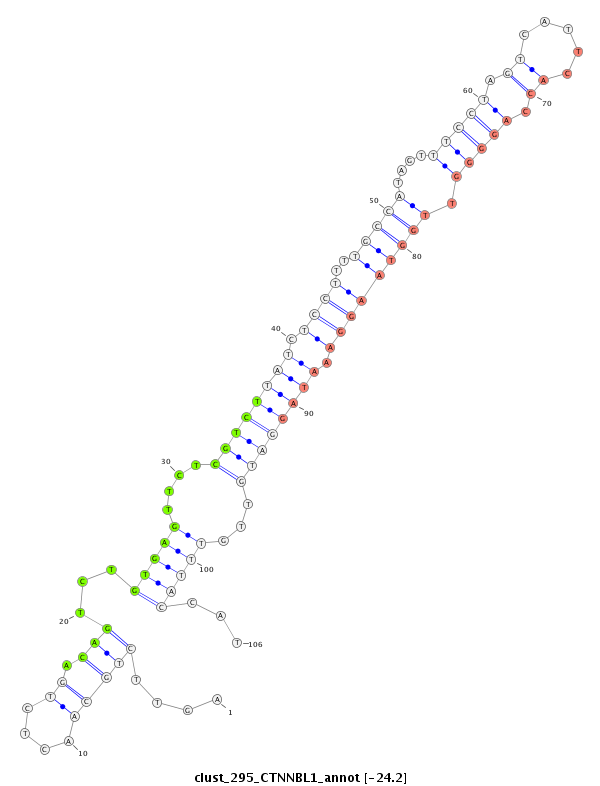
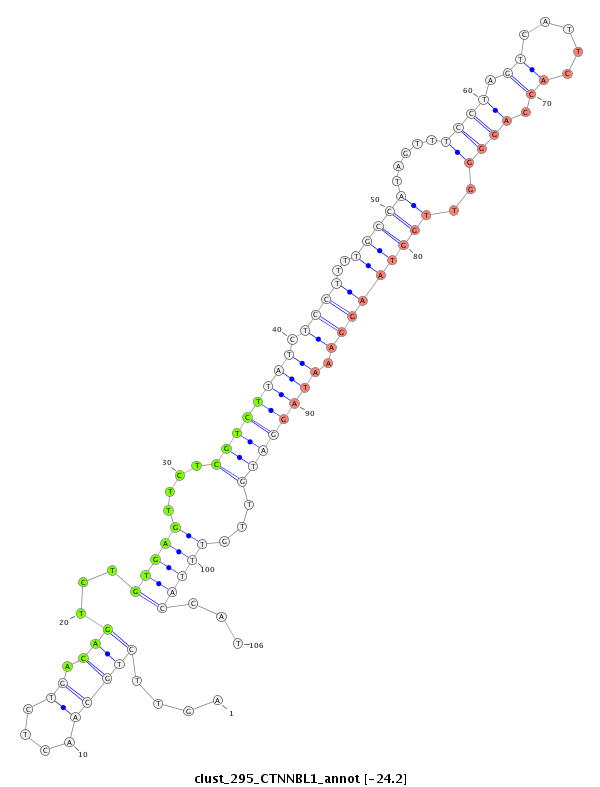
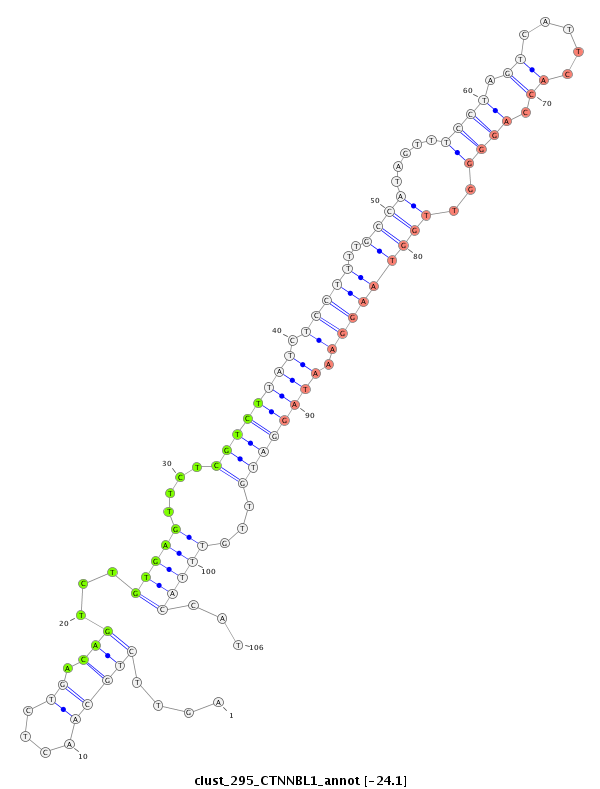
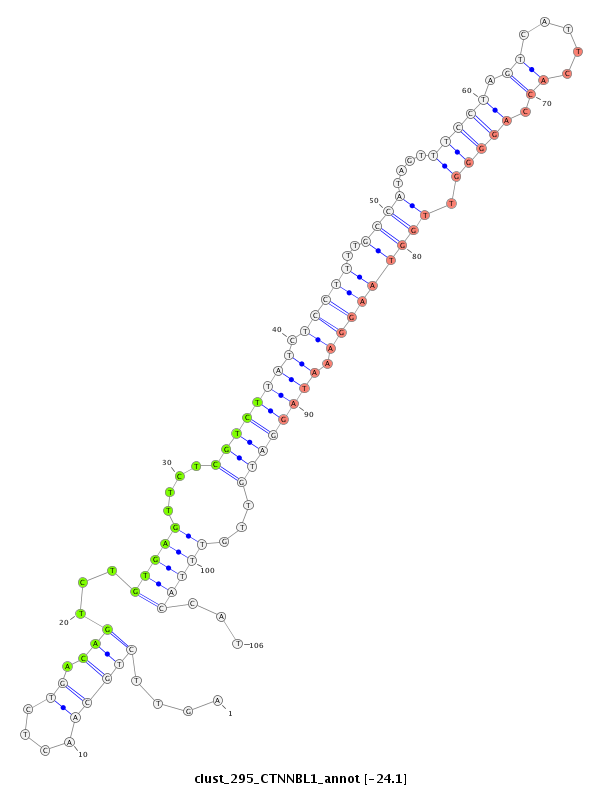
ID:clust_295_CTNNBL1 |
Coordinate:chr20:37711106-37711327 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -24.2 | -24.2 | -24.1 | -24.1 |
 |
 |
 |
 |
|
CTCATATGGTGTGCCCTACCCTAGCTGGACCTTTAAGACCCAACCTTTGGCCCTAGCCCATTATCTGGCCCATCTGATCCTCCTGTGAAAGGACAGTTTGAGCAAAACCTTGCTTGAGAAGCTTACAGACTTTCCAGTTCTGCAACTCTGACAGTCTGTGAGTTCTCGTCTTATCTCCTTTTGCCATAGTTTCCTAGTCATTCACCAGGGGTTGGTAAGGAAATAGGATGTTGTTTACCATTGTTTCTAGGGCATTGTAACATATGCTGCTAGTCAGGCTGTGAGACAATCCAGATTTTGGGGCGGTTAGCAAATTTCATTT
***************************************************************************************************************************************..................((...((((....((((((((.((((..(((((.....((((.((.....)).))))..))))))))).))))))))...))))..))********************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139208 testes |
TAX577453 breast |
RoviraIPAgo2 breast |
SRR139172 kidney |
SRR139201 placenta |
SRR139202 spleen |
SRR139204 spleen |
SRR191400 breast |
SRR191459 breast |
SRR191500 breast |
SRR191557 breast |
SRR191599 breast |
TAX577742 breast |
TAX577740 breast |
SRR342901 heart |
GSM450602 brain |
SRR039615 liver |
GSM450601 brain |
SRR037876 fibroblast |
SRR342896 heart |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................ACAGTCTGTGAGTTCTCGTCT....................................................................................................................................................... | 21 | 0 | 1 | 3.00 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................CAGTCTGTGAGTTCTCGTCTT...................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CTCATATGGTGTGC.................................................................................................................................................................................................................................................................................................................... | 14 | 0 | 8 | 1.00 | 8 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TTTGGCCCTAGCCCATTATT................................................................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................GTTCTCGTCTTATCTC................................................................................................................................................. | 16 | 0 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................ACAGTCTGTGAGTTCTCGTC........................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TCACCAGGGGTTGGTAAGGAAATAG................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................ACAGTCTGTGAGTTCTCGT......................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ATGGTGTGCCCTACC.............................................................................................................................................................................................................................................................................................................. | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....ATGGTGTGCCCTACCCTA........................................................................................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TGAGGAAGTAGGATGTTGTTTA..................................................................................... | 22 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................ACAGTCTGCGAGTTCTCTTCT....................................................................................................................................................... | 21 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................ATATGCTGCTAGT................................................ | 13 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TAAGGCAATAGGATGTCGGTTA..................................................................................... | 22 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AGGCTGTGAGACGATCCG............................. | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TGCGTGAGAAGCTTACATAGT............................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................TGAGGTAGTAGGATGTTGTTTA..................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................GTGAAAGGACAG.................................................................................................................................................................................................................................. | 12 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................................................................................................................TAAGGTAATAGGATGTT.......................................................................................... | 17 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GAGTATACCACACGGGATGGGATCGACCTGGAAATTCTGGGTTGGAAACCGGGATCGGGTAATAGACCGGGTAGACTAGGAGGACACTTTCCTGTCAAACTCGTTTTGGAACGAACTCTTCGAATGTCTGAAAGGTCAAGACGTTGAGACTGTCAGACACTCAAGAGCAGAATAGAGGAAAACGGTATCAAAGGATCAGTAAGTGGTCCCCAACCATTCCTTTATCCTACAACAAATGGTAACAAAGATCCCGTAACATTGTATACGACGATCAGTCCGACACTCTGTTAGGTCTAAAACCCCGCCAATCGTTTAAAGTAAA
*********************************************************************************..................((...((((....((((((((.((((..(((((.....((((.((.....)).))))..))))))))).))))))))...))))..))*************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139164 heart |
SRR139170 heart |
SRR139182 liver |
SRR139166 heart |
SRR139177 liver |
SRR139179 liver |
SRR139188 lung |
SRR139212 testes |
SRR139218 thymus |
SRR139219 thymus |
SRR553573 cerebellum |
TAX577740 breast |
SRR342897 heart |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................CTTCGAATGTCTGAAAG............................................................................................................................................................................................ | 17 | 0 | 2 | 5.00 | 10 | 4 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................ATTGTATACGACGAT.................................................. | 15 | 0 | 18 | 1.00 | 18 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................GGTAGACTAGGAGGAC............................................................................................................................................................................................................................................. | 16 | 0 | 4 | 1.00 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GAACGAACTCTTCGAA...................................................................................................................................................................................................... | 16 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................TCCCCAACCATTCCT..................................................................................................... | 15 | 0 | 13 | 1.00 | 13 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................GTCTAAAACCCCGCCAATCG........... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................AAAACCCCGCCAATCG........... | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................CTGTCAGACACTCAA.............................................................................................................................................................. | 15 | 0 | 12 | 1.00 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GGAAAACGGTATCAA................................................................................................................................... | 15 | 0 | 13 | 1.00 | 13 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GTCTCAAAGGTCAAGACGTCGGGA............................................................................................................................................................................. | 24 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................GTTGGTAACTGGGATCGGGCA..................................................................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................TTGGAAACCGGGA............................................................................................................................................................................................................................................................................ | 13 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/21/2015 at 07:55 PM