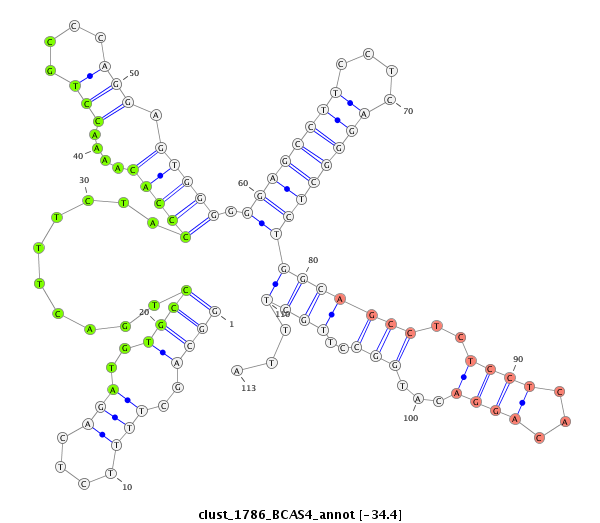
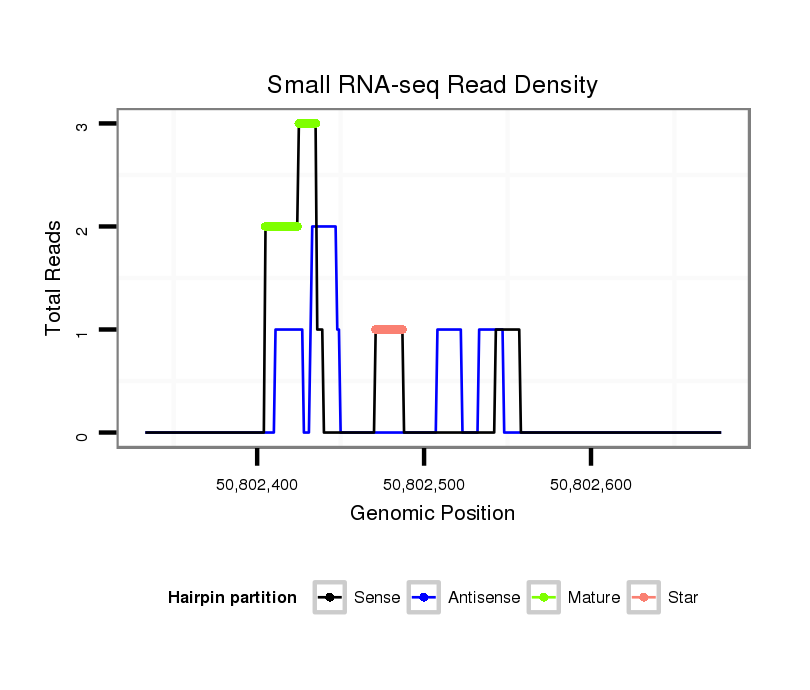
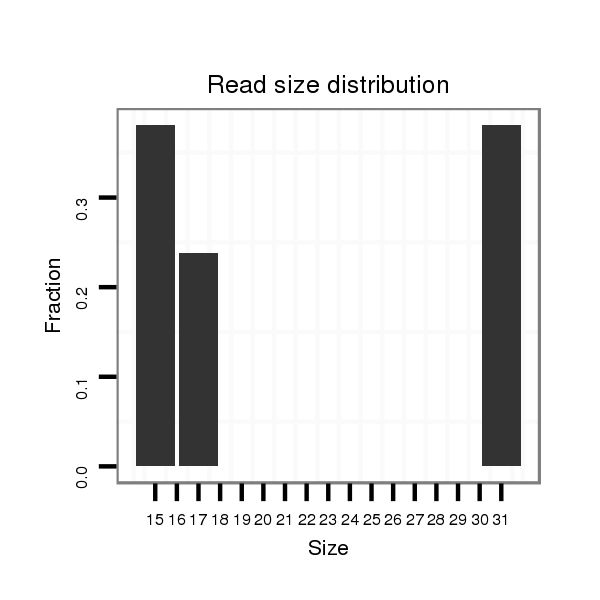
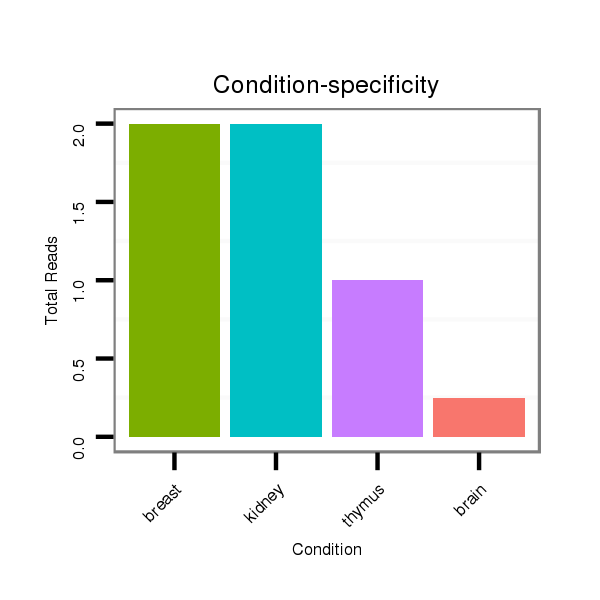
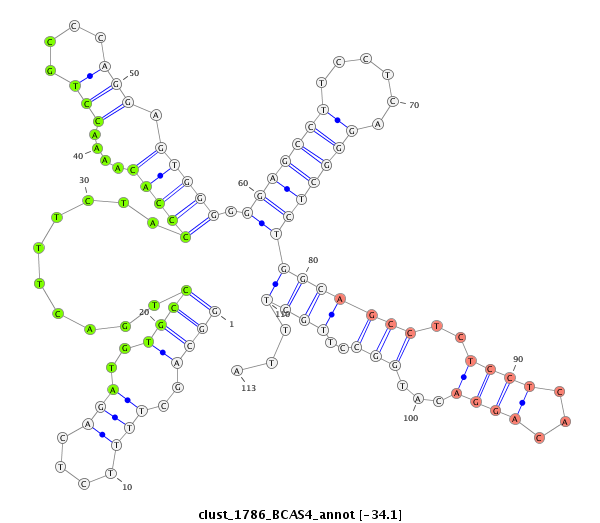
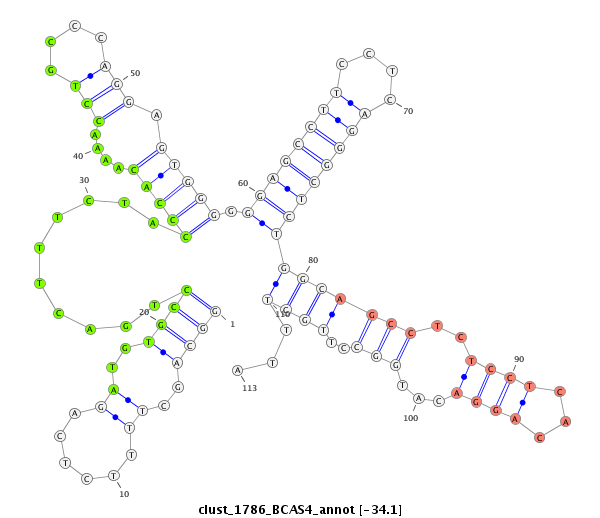
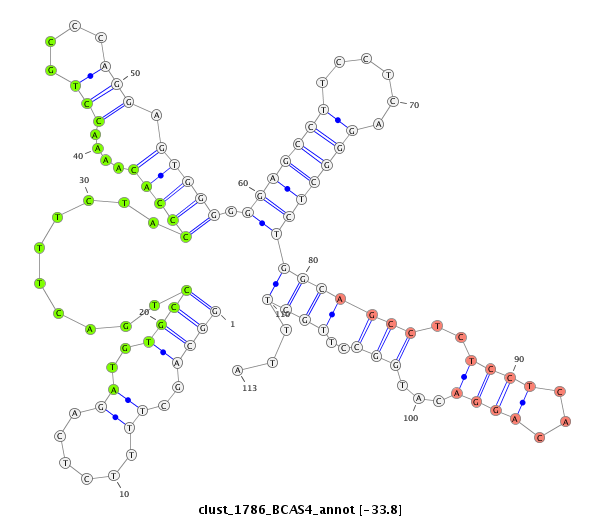
ID:clust_1786_BCAS4 |
Coordinate:chr20:50802383-50802628 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -34.1 | -34.1 | -33.8 |
 |
 |
 |
| mature | star |
|
CCTGCATCAGAACCTTCCGGGGCATGAGGAGCCAGGGGCAAGGCAGGTGCTGGGAGAGGCAGCTTTTCTCAGATGTGCCTGACTTTCTACCCACAAAACCTGCCCAGGAGTGGGGGGAGCCTTCCTCAGGGCTCTGGCAGCCTCTCCTCACAGGACATGGCCTTGCTTTAGGTGAGGGTCTGCCTAGAGCCTTGGATGAAGGTCTCCCATCCGGCCCAGCTTCAGCGTCACACAGAGGCCTTAAAACAAACACGTTAGGCCAGGCATGTTGGCCCACGCCTGTAATCCCAGCACTTTGGGAAGCCGAGGCAGGTGGATCACTTGAGGTTAGGAGTTCCAGACCAGC
*********************************************************((((..(((....)))..))))..........(((((....(((....))).))))).((((((((....))))))))(((((((..((((...))))...)))..))))...******************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | RoviraIPAgo2 breast |
SRR139173 kidney |
SRR139176 kidney |
SRR139219 thymus |
SRR553574 heart |
TAX577740 breast |
GSM450605 brain |
SRR037876 fibroblast |
SRR330914 skin |
SRR342900 heart |
GSM450606 brain |
SRR039623 liver |
SRR191422 breast |
SRR330915 skin |
SRR342895 heart |
TAX577743 breast |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................ATGTGCCTGACTTTCTACCCACAAAACCTGC................................................................................................................................................................................................................................................... | 31 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........AACCTGCCGGGGCATGAG.............................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGCCTCTCCTCACAGGA............................................................................................................................................................................................... | 17 | 0 | 3 | 1.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................ACAAAACCTGCCCAG............................................................................................................................................................................................................................................... | 15 | 0 | 17 | 1.00 | 17 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CCGGCCCAGCTTCAG......................................................................................................................... | 15 | 0 | 7 | 1.00 | 7 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................ACGTTAGGCCCCGAATGTTGG.......................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CACAGGACATGGCCTTG..................................................................................................................................................................................... | 17 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AGGTATCCCGTCCGGCCCATC.............................................................................................................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................CGAGGCATGTCGGCCCACGGC.................................................................. | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GAACCTGCCCAGGAGAGGGGG...................................................................................................................................................................................................................................... | 21 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................CGAGGGAGCTTTTCTCATATGT.............................................................................................................................................................................................................................................................................. | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............CCTTCCGGGG.................................................................................................................................................................................................................................................................................................................................... | 10 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................ATCGGGGGGAGCCTTCCT............................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................................................................................CAGCACTTTGGGAAGCCGAGG..................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................GGCAGGTGGATCACTTGAGG................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................TAGGAGTTCC........ | 10 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GGACGTAGTCTTGGAAGGCCCCGTACTCCTCGGTCCCCGTTCCGTCCACGACCCTCTCCGTCGAAAAGAGTCTACACGGACTGAAAGATGGGTGTTTTGGACGGGTCCTCACCCCCCTCGGAAGGAGTCCCGAGACCGTCGGAGAGGAGTGTCCTGTACCGGAACGAAATCCACTCCCAGACGGATCTCGGAACCTACTTCCAGAGGGTAGGCCGGGTCGAAGTCGCAGTGTGTCTCCGGAATTTTGTTTGTGCAATCCGGTCCGTACAACCGGGTGCGGACATTAGGGTCGTGAAACCCTTCGGCTCCGTCCACCTAGTGAACTCCAATCCTCAAGGTCTGGTCG
********************************************************************************************************************************************************************************((((..(((....)))..))))..........(((((....(((....))).))))).((((((((....))))))))(((((((..((((...))))...)))..))))...********************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR139207 spleen |
SRR139188 lung |
SRR139193 ovary |
SRR330907 skin |
SRR330916 skin |
SRR553572 frontal-cortex |
SRR330919 skin |
SRR330917 skin |
SRR330921 skin |
GSM450602 brain |
SRR039611 liver |
SRR342895 heart |
SRR342898 heart |
SRR444050 skin |
TAX577744 breast |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................ACGGGTCCTCACCCC....................................................................................................................................................................................................................................... | 15 | 0 | 20 | 1.10 | 22 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................CCCAGACGGATCTCG............................................................................................................................................................ | 15 | 0 | 5 | 1.00 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GACTGAAAGATGGGTGT........................................................................................................................................................................................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GACGGGTCCTCACCCCCC..................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CCAGAGGGTAGGCCG................................................................................................................................... | 15 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CCAGAGGGTAGG...................................................................................................................................... | 12 | 0 | 20 | 0.40 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................CCCTCTCCGTCGAAAAG...................................................................................................................................................................................................................................................................................... | 17 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................GCCAGAGGGTAGGCC.................................................................................................................................... | 15 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GTGCGGACATTAGGGTCGTGAAAC................................................ | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................ACACCCGGGGGCGGACATCAGGGTCGTGA................................................... | 29 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....TATTCTTGGTAGTCCCCGTACT............................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................CCCTCGGAAGTAGTCCC....................................................................................................................................................................................................................... | 17 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................ATCCTCAAGGTCTGGTC. | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/21/2015 at 08:21 PM