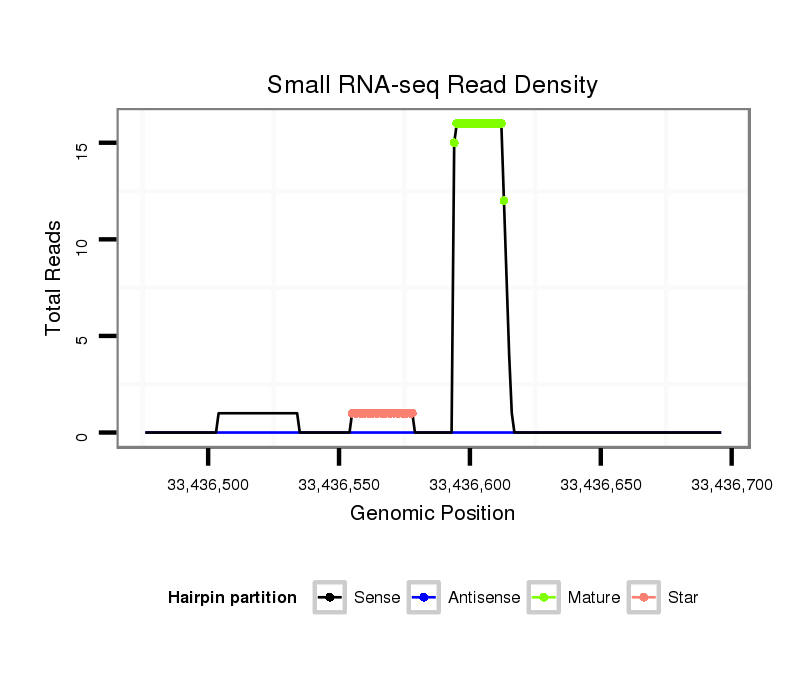
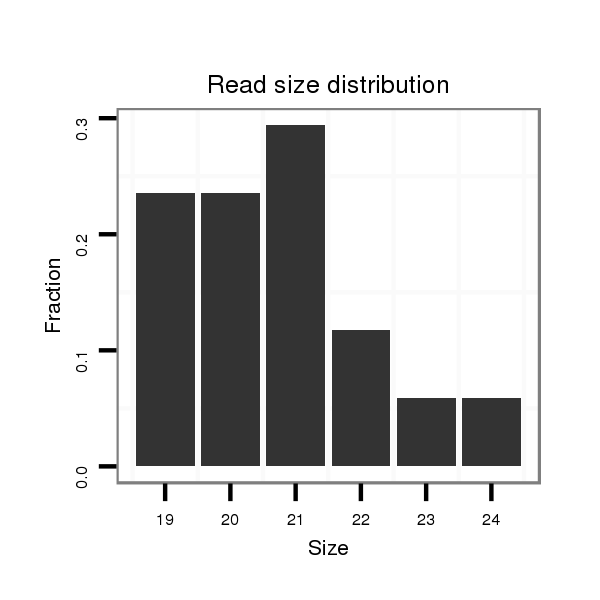
| ......................................................................................................................CTTGGACCTCTGATGGGAGC................................................................................... |
20 |
0 |
1 |
4.00 |
4 |
4 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGCC.................................................................................. |
21 |
0 |
1 |
4.00 |
4 |
4 |
| ......................................................................................................................CTTGGACCTCTGATGGGAG.................................................................................... |
19 |
0 |
1 |
4.00 |
4 |
4 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGCCC................................................................................. |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGCCCTAAT............................................................................. |
26 |
3 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGA................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................GTTGGACCTCTGATGGGAGCCCAAAT............................................................................. |
26 |
3 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGT................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................TTGGACCTCTGATGGGAGCCCTA............................................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................TTGGACCTCTGATGGGAGCCCATG.............................................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGCCCGAAA............................................................................. |
26 |
2 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................TTGGACCTCTGATGGGAGCCC................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................CATGGACCTCTGATGGGAGCCCA................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................TTGGACCTCTGATGGGAGCCCAAT.............................................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................AGCCCCTTTAAGAGGTCCCCAGCC...................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
| ............................TACCAAGGAGTCACTCCCAACCCTGCAGGGC.................................................................................................................................................................. |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................CTTGGACCTCTGATGGGAGCCCA................................................................................ |
23 |
0 |
1 |
1.00 |
1 |
1 |