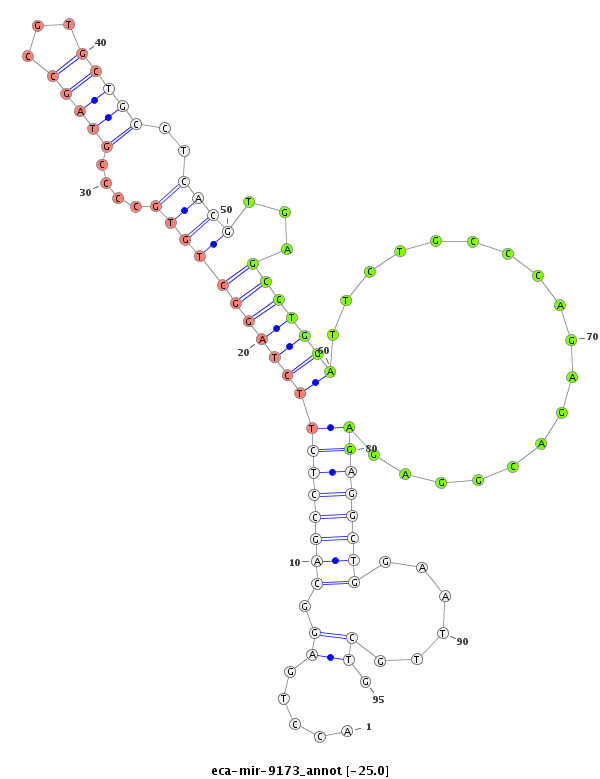
ID:eca-mir-9173 |
Coordinate:chr16:1841484-1841626 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
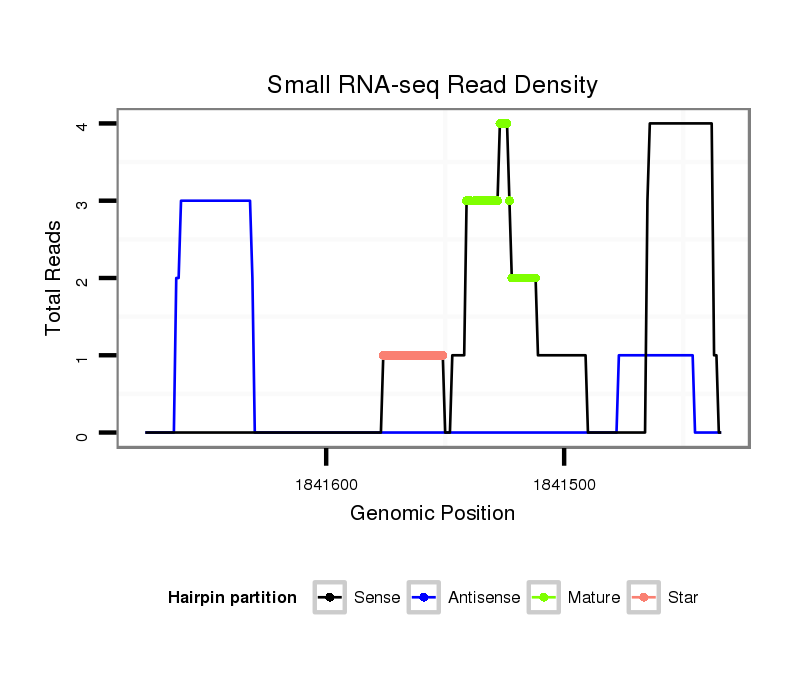
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -33.0 | -32.4 | -32.4 | -32.3 |
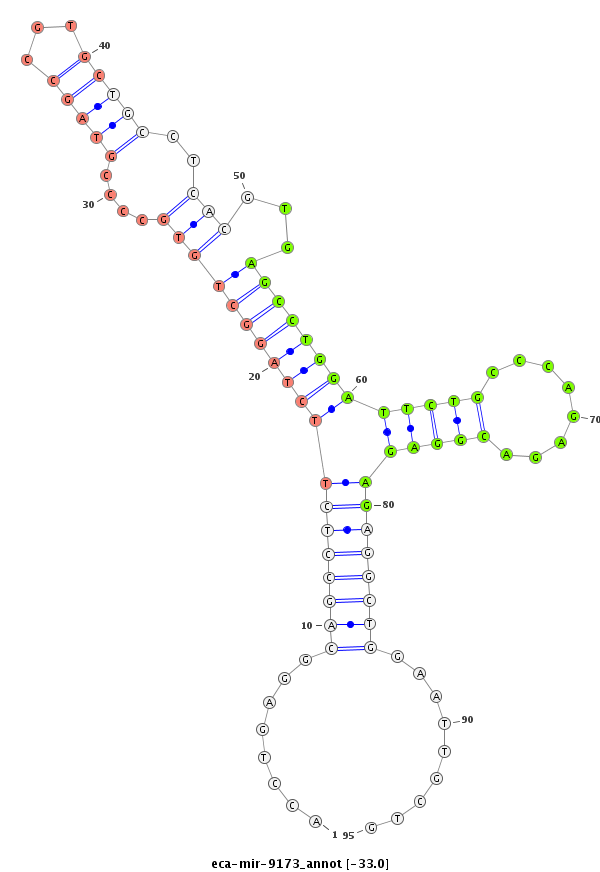 |
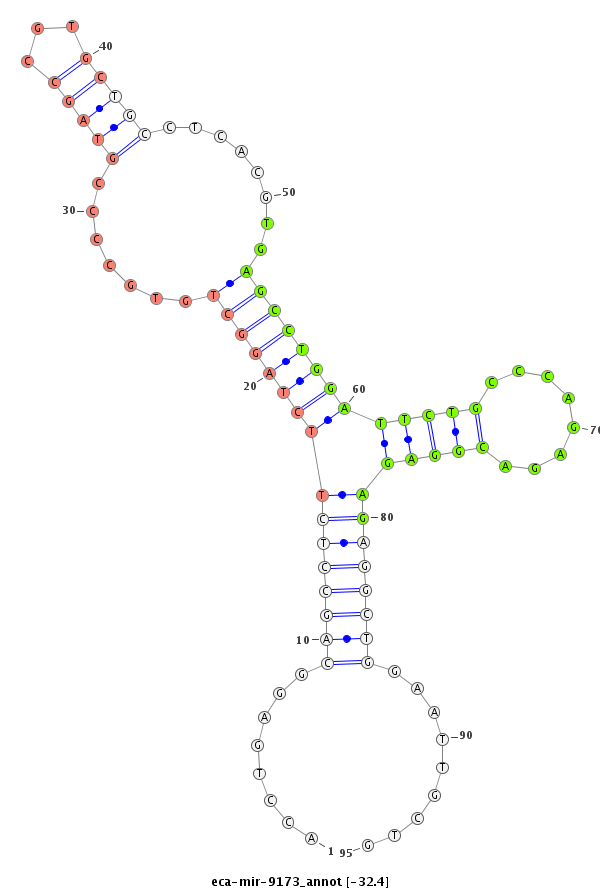 |
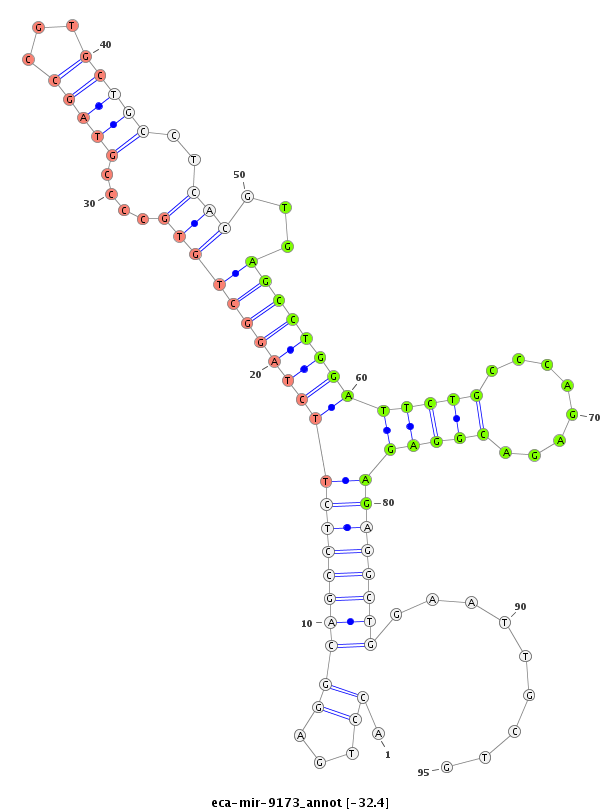 |
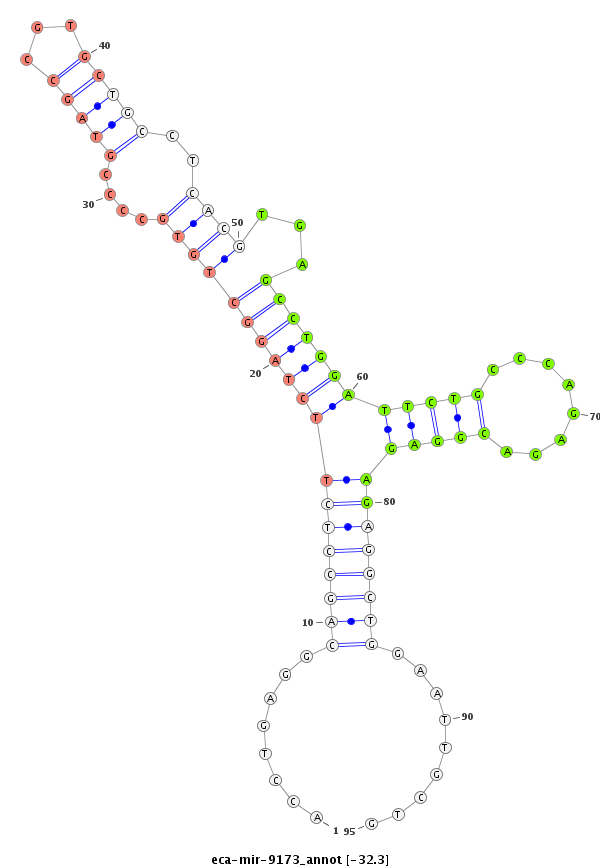 |
|
ACTCTTTTTCCAGAAGGTGGAAAACTTGGGGTTCTTACAAGTGGAACCATTTCTTCCCAGTCTTCACCTGAGGGGCTTTCTTCTGACCTGAGGCAGCCTCTTCTAGGCTGTGCCCCGTAGCCGTGCTGCCTCACGTGAGCCTGGATTCTGCCCAGAGACGGAGAGAGGCTGGAATTGCTGAGACCTGGGAGTGCAGGATATTCTGCTGGTTTAGGAGACTTCACAGTGGAAGCTGGCGCTTGC
*************************************************************************************.....((.(((((((((((((((((((....(((((...)))))..))))...)))))))..................))))))))......)).*************************************************************** |
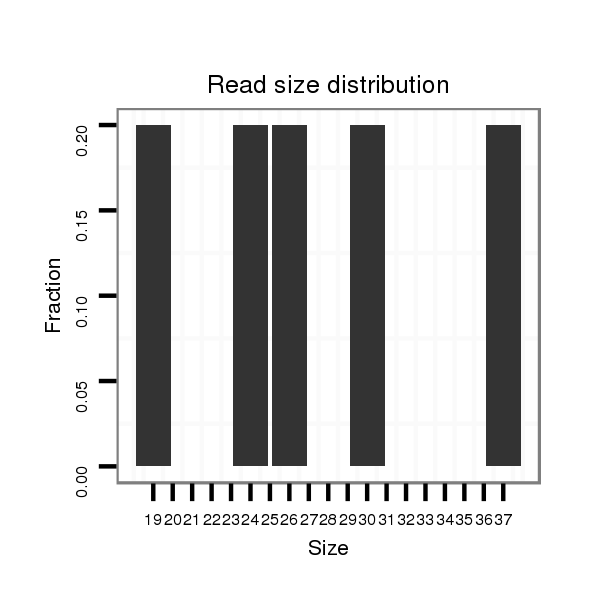
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................TAGGAGACTTCACAGTGGAAGCTGGCGC.... | 28 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................................................................TGAGCCTGGATTCTGCCCAGAGACGGAGAG.............................................................................. | 30 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................................................................................TGAGCCTGGATTCTGCCCA......................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
| .................................................................................................................................CTCACGTGAGCCTGGATTCTGCCC.......................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
| ..............................................................TTCACCTGAGGGGCTTTCTTCTGAAT........................................................................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 |
| ...................................................................................................................................................................................................................TAGGAGACTTCACAGTGGAAGCTGGCGCTT.. | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................................................................................................................................AGGAGACTTCACAGTGGAAGCTGGCGC.... | 27 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................................................................................................................GCCCAGAGACGGAGAGAGGCTGGAATTGCTGAGACCT......................................................... | 37 | 0 | 1 | 1.00 | 1 | 1 |
| ...................................................................................................................................................................................................................TAGGAGACTTCACAGTGGAAGCTGGCT..... | 27 | 1 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................TTCTAGGCTGTGCCCCGTAGCCGTGC..................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
|
TGAGAAAAAGGTCTTCCACCTTTTGAACCCCAAGAATGTTCACCTTGGTAAAGAAGGGTCAGAAGTGGACTCCCCGAAAGAAGACTGGACTCCGTCGGAGAAGATCCGACACGGGGCATCGGCACGACGGAGTGCACTCGGACCTAAGACGGGTCTCTGCCTCTCTCCGACCTTAACGACTCTGGACCCTCACGTCCTATAAGACGACCAAATCCTCTGAAGTGTCACCTTCGACCGCGAACG
***************************************************************.....((.(((((((((((((((((((....(((((...)))))..))))...)))))))..................))))))))......)).************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .............TTCCACCTTTTGAACCCCAAGAATGTTCACCTT..................................................................................................................................................................................................... | 33 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................................................................................................................................TAAGACGACCAAATCCTCTGAAGTGTCACCTT............ | 32 | 0 | 1 | 1.00 | 1 | 1 |
| ............................TTCAAGAATGTTCACCTTGGTAAAGAAGGGT........................................................................................................................................................................................ | 31 | 2 | 1 | 1.00 | 1 | 1 |
| ......TAAGGTCCTCCACCTTTTGAACCCCAAGAAT.............................................................................................................................................................................................................. | 31 | 2 | 1 | 1.00 | 1 | 1 |
| ...............CCACCTTTTGAACCCCAAGAATGTTCACCT...................................................................................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:43 AM