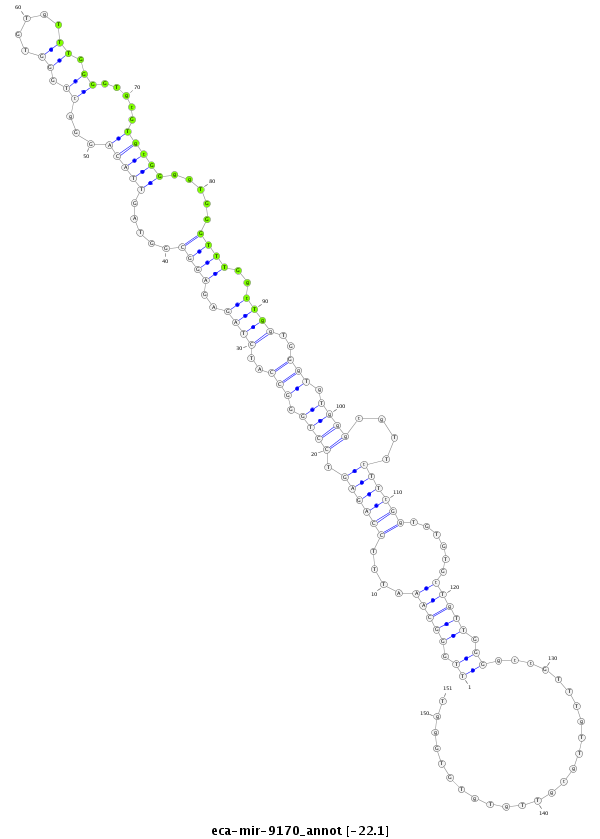



ID:eca-mir-9170 |
Coordinate:chr16:18046864-18046984 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
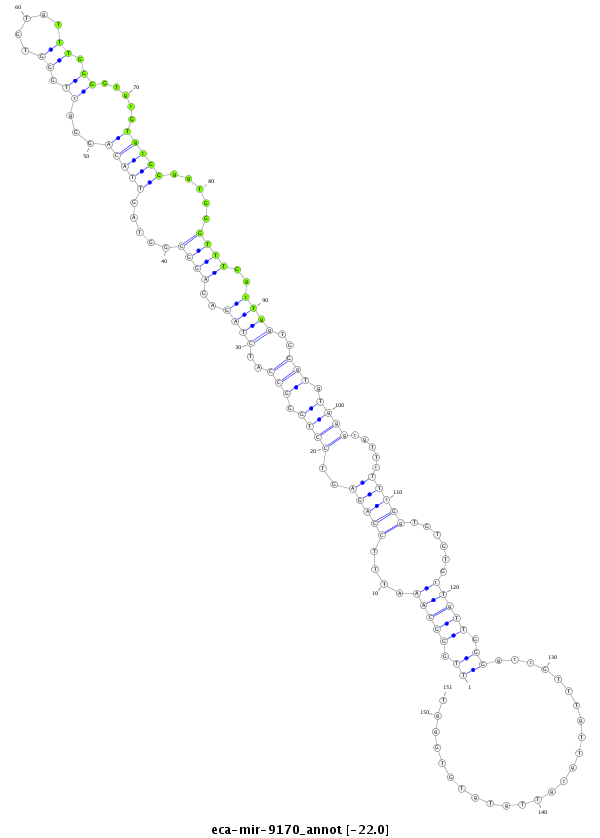
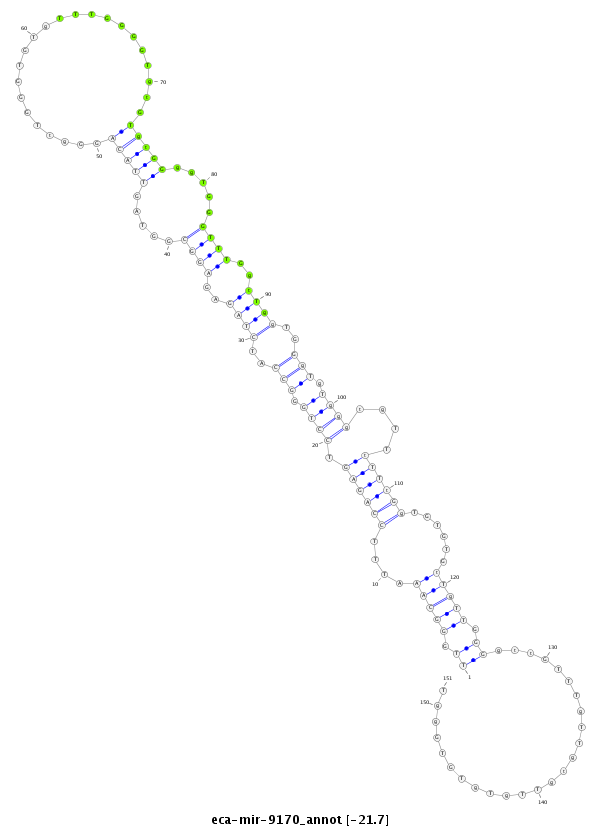
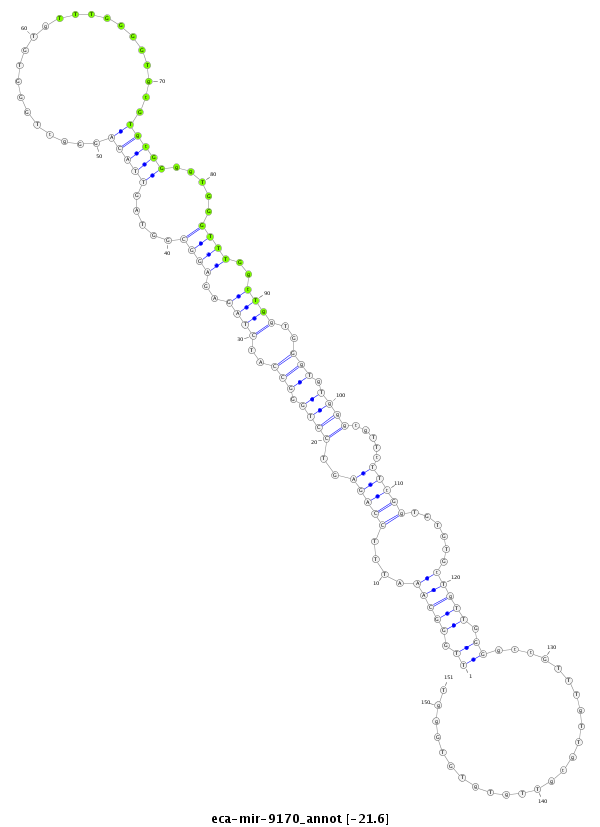
| -22.0 | -21.7 | -21.6 |
 |
 |
 |
|
CCACGTTTTCTTATCTTCTCTATCCACAGCATGACTTGGGCAAATTTCCAGAGTCCTGGGCCATCTAGAGAGGCGGTAGTTACAGGgtTGGGTGTgTTTGGGGTgtGTgtGGggTGGGTTTGgtTggTGGgTgTgggtgTTtTTtGgTGTGTGtTgTTGGGgttGTTTgTTgtgTTgTgTGTGggTgtGTTTtgTgtTgTtTttttTGGTGtgtGgtggTG
***********************************((.(((((....((((((.((((.(((..((((..((((.....(((((...((.((.....)).)).....))))).....))))..))))..))).))))....))))))......))))).)).........................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .............................................................................................................................................................TGGGCAAGTTTCTTCACTTCTCTGTGCCT................................... | 29 | 0 | 1 | 22.00 | 22 | 22 |
| .............................................................................................................................................................TGGGCAAGTTTCTTCACTTCTCTGTGCC.................................... | 28 | 0 | 1 | 7.00 | 7 | 7 |
| .....................................................................................................................................................TGTGATCTTGGGCAAGTTTCTTCACTTCTC.......................................... | 30 | 0 | 1 | 3.00 | 3 | 3 |
| .................................................................................................................................................................................................CTCATCTATAAAATGGTGACAGCACCTG | 28 | 0 | 1 | 2.00 | 2 | 2 |
| ........................................................................GCGGTAGTTACAGGCATGGGTGTCTTTGGT....................................................................................................................... | 30 | 1 | 1 | 2.00 | 2 | 2 |
| ...............................TGACTTGGGCAAATTTCCAGAGTCCTGGGCCT.............................................................................................................................................................. | 32 | 1 | 1 | 2.00 | 2 | 2 |
| ..........................................................................................................................................................TCTTGGGCAAGTTTCTTCACTTCTCTGTGCCT................................... | 32 | 0 | 1 | 2.00 | 2 | 2 |
| ...............................................................TCTAGAGAGGCGGTAGTTACAGGCATGGA................................................................................................................................. | 29 | 1 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................................................................TGGGCAAGTTTCTTCACTTCTGTGTGCCT................................... | 29 | 1 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................................................................TAGCTGTGTGATCTTGGGCAAGTTTCTTCACT.............................................. | 32 | 0 | 1 | 1.00 | 1 | 1 |
| ...............TTCTCTATCCACAGCATGACTTGGGC.................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ................................................................................................TTTGGGGTCAGTCAGGCCTGGGTTTGCATC............................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| .................................................................TAGAGAGGCGGTAGTTACAGGCATGGGTTTTTT........................................................................................................................... | 33 | 2 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................................................................................................TTTACTCATCTATAAAATGGTGACAGCACCTG | 32 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................TGACTTGGGCAAATTTCCAGAGTCCTGGGCA............................................................................................................................................................... | 31 | 1 | 1 | 1.00 | 1 | 1 |
| ......TTTCTTATCTTCTCTATCCACAGCATGACT......................................................................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................TGACTTGGGCAAATTTCCAGAGTCCTGGGC................................................................................................................................................................ | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ................................................................................................................................................AGCTGTGTGATCTTGGGCAAGTTTCTTCACT.............................................. | 31 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................................................................................................................TGTGATCTTGGGCAAGTTTCTTCACTTCTCT......................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................TTTGCATCCTGGCTCTCCCACTTGTTAGCTG........................................................................ | 31 | 1 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................................................................................................................ATCTATAAAATGGTGACAGCAGCTG | 25 | 1 | 1 | 1.00 | 1 | 1 |
| ........TCTTATCTTCTCTATCCACAGCATGACTTGT...................................................................................................................................................................................... | 31 | 1 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................................................................TGGGCAAGTTTCTTCACTTTTCTGTGCCT................................... | 29 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................GTGATCTTGGGCAAGTTTCTTCACTTC............................................ | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................GTGATCTTGGGCAAGTTTCTTCACTTCTCTGT....................................... | 32 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................................................................TGGGCAAGTTTCTTCACTTCTCTGTGCCG................................... | 29 | 1 | 1 | 1.00 | 1 | 1 |
| ............................................................................................TGACTTTGGGGTCGGTCAGGCCTGGGTTT.................................................................................................... | 29 | 2 | 2 | 0.50 | 1 | 1 |
| .....................................................................................................................................................TGTGATCTTGGGCAAGTTTCTTCACTTCATT......................................... | 31 | 2 | 2 | 0.50 | 1 | 1 |
|
GGTGCAAAAGAATAGAAGAGATAGGTGTCGTACTGAACCCGTTTAAAGGTCTCAGGACCCGGTAGATCTCTCCGCCATCAATGTCCgtACCCACAgAAACCCCAgtCAgtCCggACCCAAACgtAggACCgAgAgggtgAAtAAtCgACACACtAgAACCCgttCAAAgAAgtgAAgAgACACggAgtCAAAtgAgtAgAtAttttACCACtgtCgtggAC
***********************************((.(((((....((((((.((((.(((..((((..((((.....(((((...((.((.....)).)).....))))).....))))..))))..))).))))....))))))......))))).)).........................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................AGAGACACGGAGTCAAATGAGTAGATAT.................. | 28 | 0 | 2 | 0.50 | 1 | 1 |
| ...............................................................................................................................................................................AGAGACACGGAGTCAAATGAGTAGATAAT................. | 29 | 1 | 4 | 0.50 | 2 | 2 |
| .................................................................................................................................................................................AGACACGGAGTCAAATGAGTAGATAT.................. | 26 | 0 | 4 | 0.25 | 1 | 1 |
| ...............................................................................................................................................................................AGAGACACGGAGTCAAATGAGTAGA..................... | 25 | 0 | 15 | 0.20 | 3 | 3 |
| ...............................................................................................................................................................................AGAGACACGGAGTCAAATGAGT........................ | 22 | 0 | 20 | 0.10 | 2 | 2 |
| ...............................................................................................................................................................................AGAGACACGGAGTCAAATGAGTAG...................... | 24 | 0 | 16 | 0.06 | 1 | 1 |
| .........................................................................................................................................................................AAATGAAGAGACACGGAGTCAAAAGAGTAGATAT.................. | 34 | 2 | 18 | 0.06 | 1 | 1 |
| ..............................................................................................................................................................................AAGAGACACGGAGTCAAATGAGTAGACATT................. | 30 | 1 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 11:44 AM