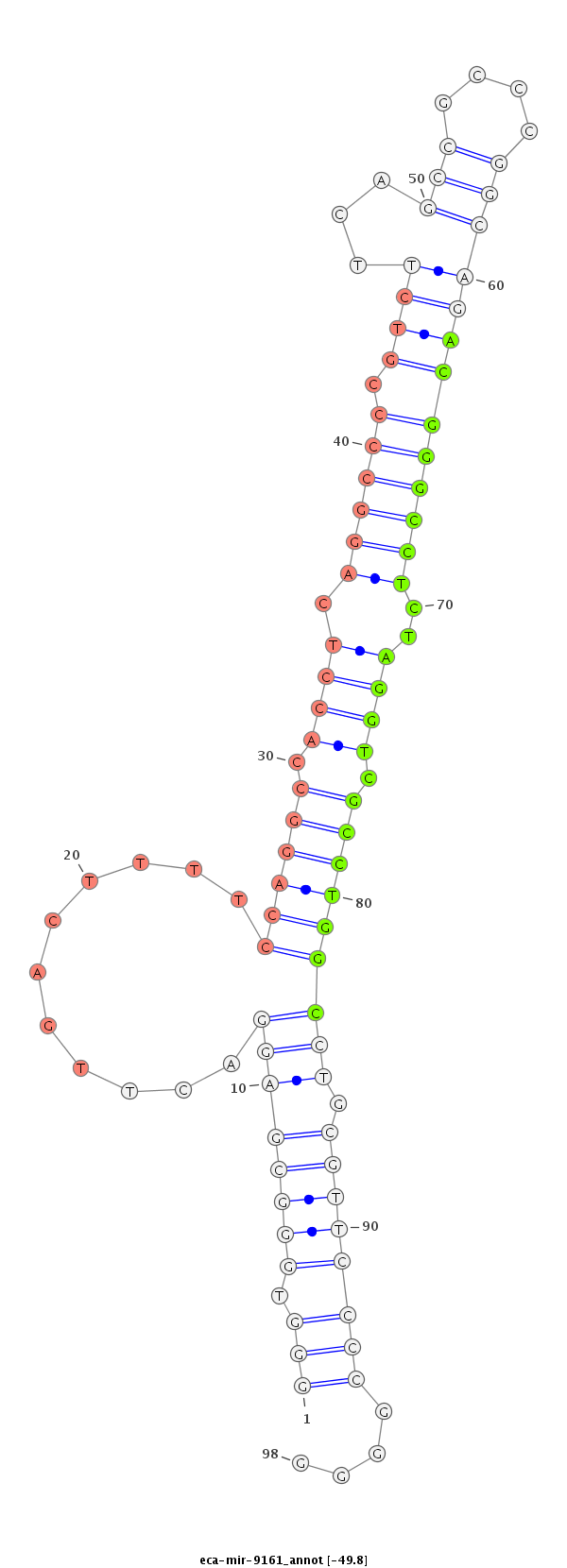
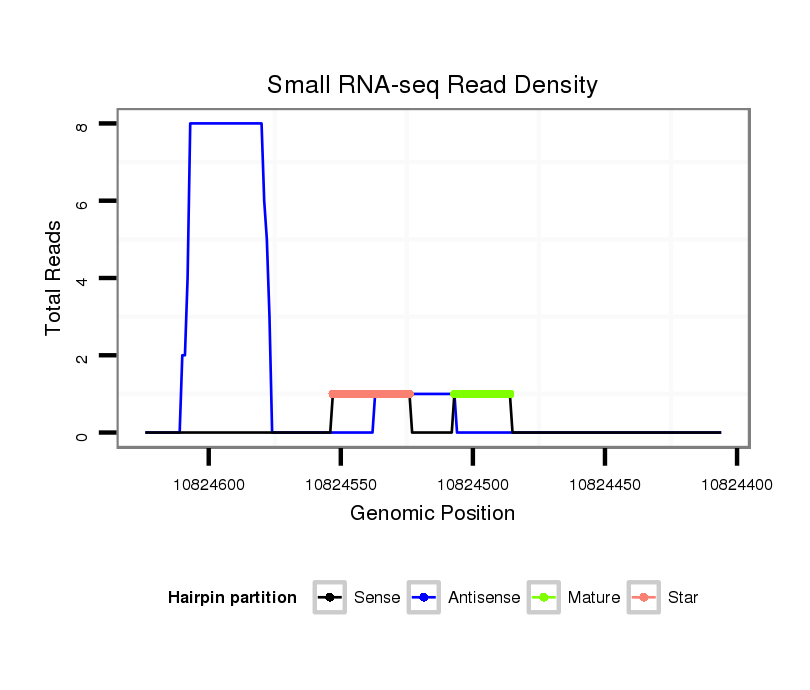
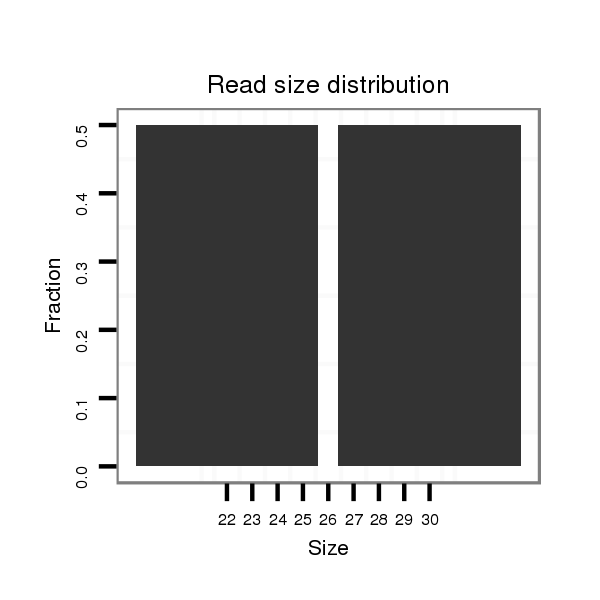
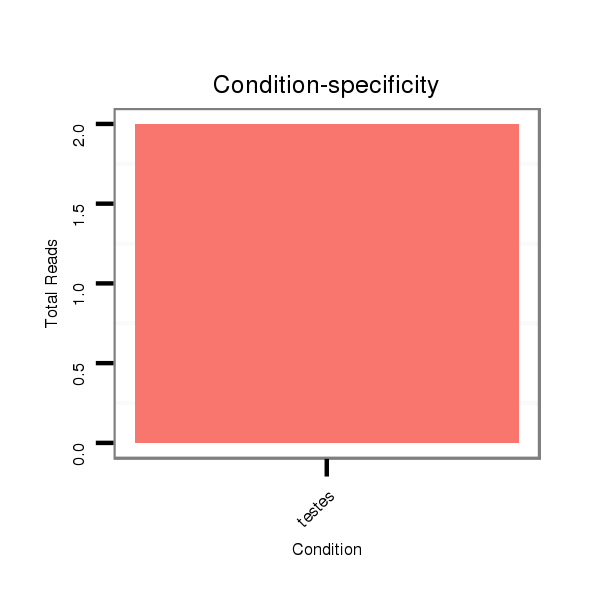
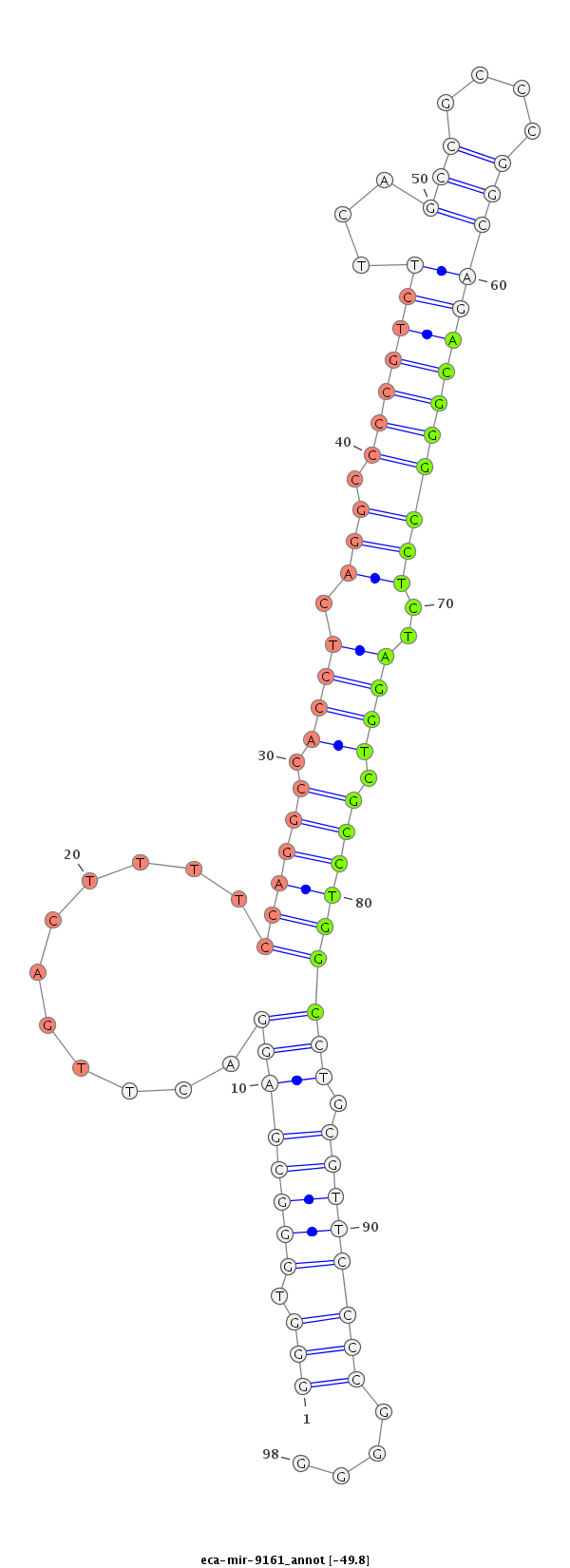
ID:eca-mir-9161 |
Coordinate:chr14:10824456-10824574 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -49.8 | -49.8 | -49.8 |
 |
 |
 |
|
TGTGGGGTCGAAGGCAGGCCAAGGTTACTCGGTTTTAAATGATCCTAATGCGGTTGGGGTGGGCGAGGACTTGACTTTTCCAGGCCACCTCAGGCCCCGTCTTCAGCCGCCCGGCAGACGGGCCTCTAGGTCGCCTGGCCTGCGTTCCCCGGGGCCGGGGCTGCTCATGGCGTCGGTAGGGCCCGGCCGTTTGGGTCAGGCGGCGAGAGGGCGGTTGGC
********************************************************(((.((((((((...........((((((.((((.((((((.((((...(((....)))))))))))))..)))).))))))))).))))))))....***************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .....................................................................................................................ACGGGCCTCTAGGTCGCCTGGC................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................TGACTTTTCCAGGCCACCTCAGGCCCCGTC...................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
|
ACACCCCAGCTTCCGTCCGGTTCCAATGAGCCAAAATTTACTAGGATTACGCCAACCCCACCCGCTCCTGAACTGAAAAGGTCCGGTGGAGTCCGGGGCAGAAGTCGGCGGGCCGTCTGCCCGGAGATCCAGCGGACCGGACGCAAGGGGCCCCGGCCCCGACGAGTACCGCAGCCATCCCGGGCCGGCAAACCCAGTCCGCCGCTCTCCCGCCAACCG
*****************************************************************(((.((((((((...........((((((.((((.((((((.((((...(((....)))))))))))))..)))).))))))))).))))))))....******************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ......................................................................................................................................................CCCCGGCCCCGACGAGTACCGCAGGCAT......................................... | 28 | 1 | 1 | 4.00 | 4 | 4 |
| .................CGGTTCCAATGAGCCAAAATTTACTAGGATT........................................................................................................................................................................... | 31 | 0 | 1 | 3.00 | 3 | 3 |
| .......................................................................................................................................................CCCGGCCCCGACGAGTACCGCAGGCAT......................................... | 27 | 1 | 1 | 2.00 | 2 | 2 |
| ..............GTCCGGTTCCAATGAGCCAAAATTTACTAGG.............................................................................................................................................................................. | 31 | 0 | 1 | 2.00 | 2 | 2 |
| .................CGGTTCCAATGAGCCAAAATTTACTAGGAT............................................................................................................................................................................ | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ................CCGGTTCCAATGAGCCAAAATTTACTAGGA............................................................................................................................................................................. | 30 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................................................................CGGGCCAGAAGTCGGCGCTCCGTCT..................................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 1 |
| ................CCGGTTCCAATGAGCCAAAATTTACTAGGAT............................................................................................................................................................................ | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................TCCCGGCCCCGACGAGTACCGCAGGCAT......................................... | 28 | 2 | 1 | 1.00 | 1 | 1 |
| .............TGTCCGGTTCCAATGAGCCAAAATTTACTAGG.............................................................................................................................................................................. | 32 | 1 | 1 | 1.00 | 1 | 1 |
| .......................................................................................GGAGTCCGGGGCAGAAGTCGGCGGGCCGTCT..................................................................................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................................................................GGCCCCGGCCCCGACGAGTACCGCAGGCAT......................................... | 30 | 1 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:40 AM