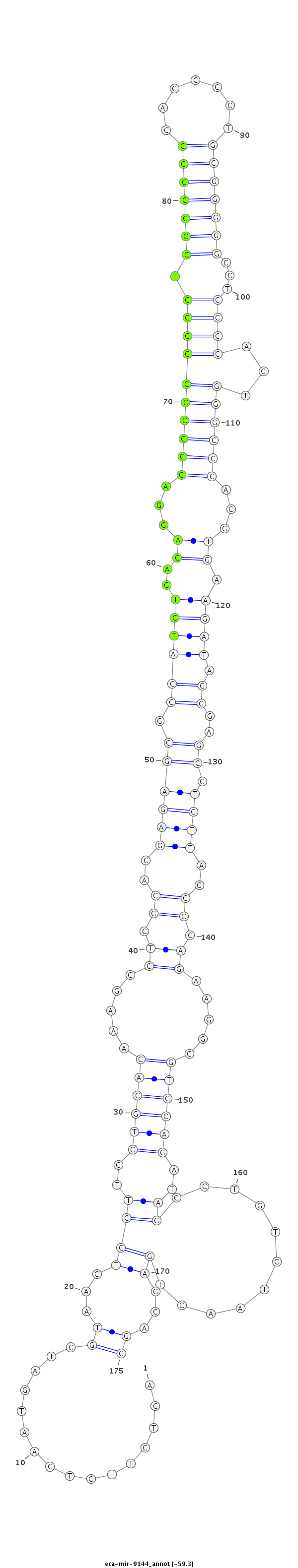
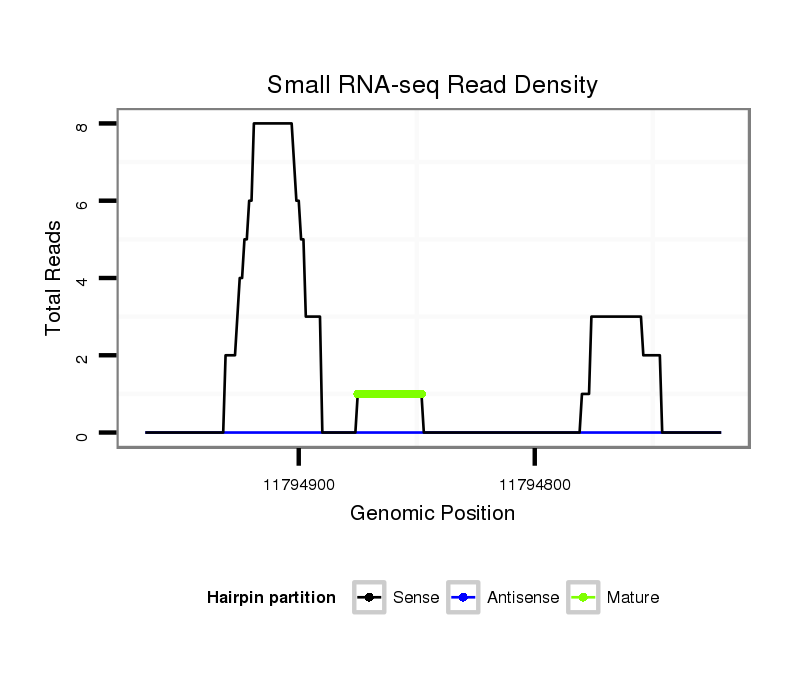
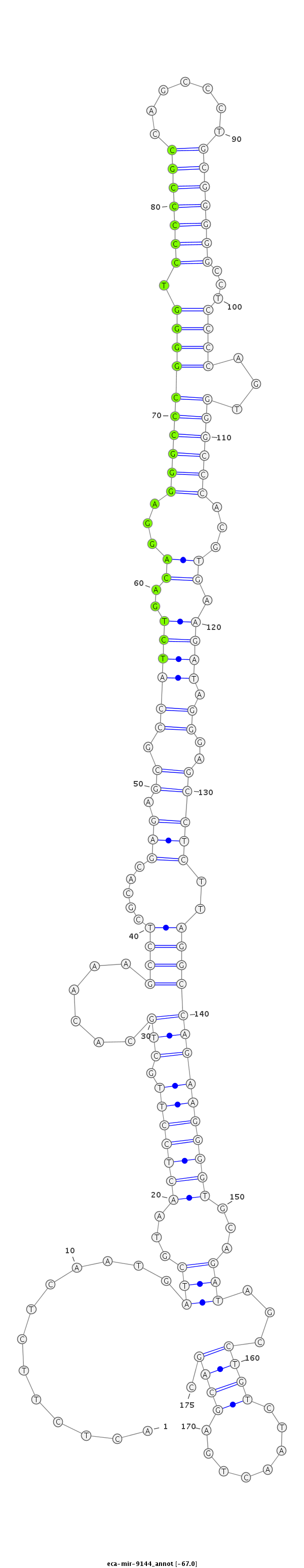
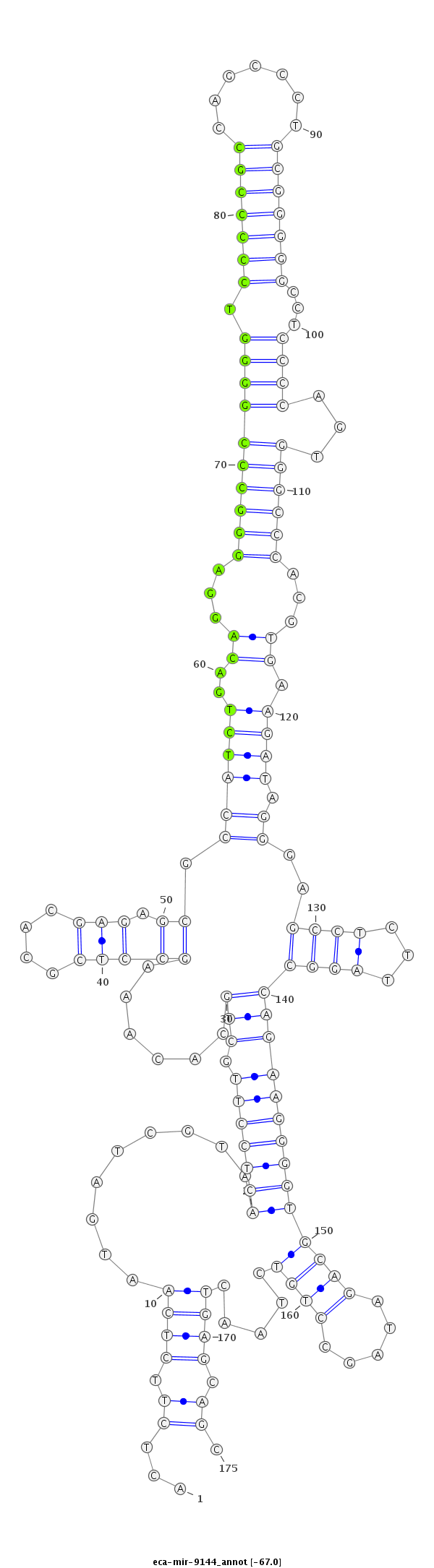
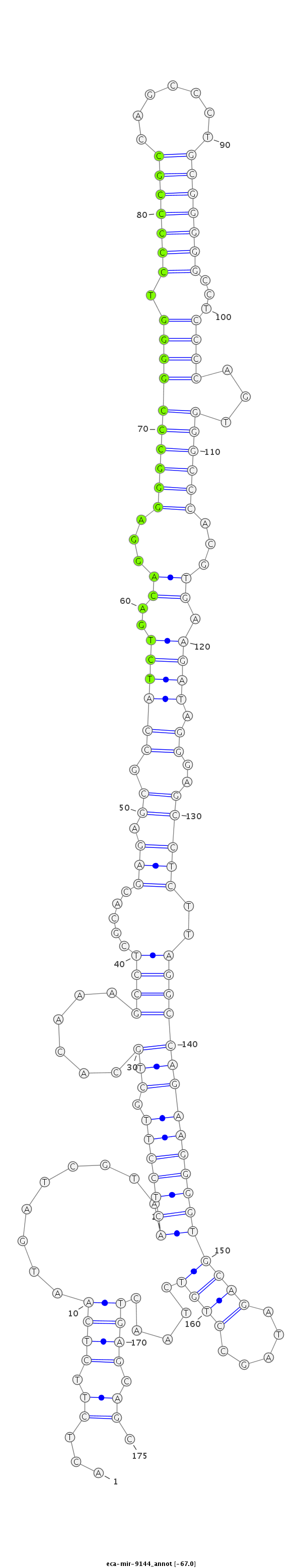
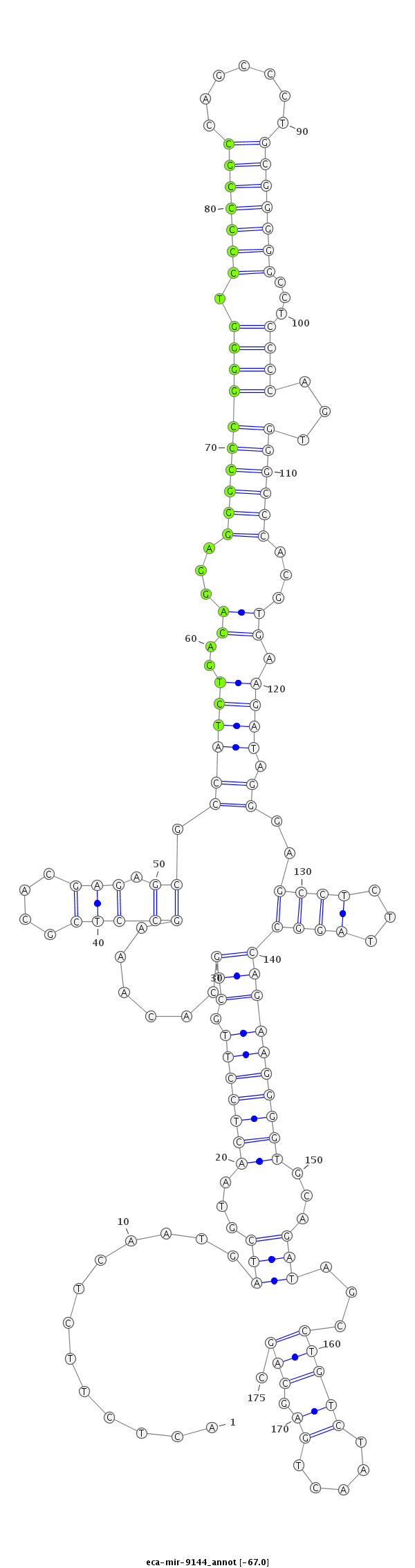
| .............................................................................................................................................................................................TAGCCTGTCTAACTGAGCAGCCAGATGGCC.......................... |
30 |
0 |
1 |
2.00 |
2 |
2 |
| ..............................................TGATCGTAACTCCTTGCTGCACAAAGCCT.......................................................................................................................................................................... |
29 |
0 |
1 |
2.00 |
2 |
2 |
| ...........................................................TTGCTGCACAAAGCCTCGCACGAGAA................................................................................................................................................................ |
26 |
1 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................TCTGACAGGAGGGCCCGGGGTCCCCCGCCT............................................................................................................................. |
30 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................TACTCTTCTCAATGATCGTAACTCCTTGC...................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................................................................................................................................................CCTGTCTAACTGAGCAGCCAGATGGCCTCCATT.................... |
33 |
2 |
1 |
1.00 |
1 |
1 |
| ..................................TACTCTTCTCAATGATCGTAACTCCTTGCTGC................................................................................................................................................................................... |
32 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................TCAATGATCGTAACTCCTTGCTGCAC................................................................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................................................................................................CAGATAGCCTGTCTAACTGAGCAGCC.................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ........................................TCTCAATGATCGTAACTCCTTGCT..................................................................................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................................................................................................CAGATAGCCTGTCTAACTGAGCAGCT.................................. |
26 |
1 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................TCTGACAGGAGGGCCCGGGGTCCCCCGC............................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................................................TCGCACGAGAGCGCCATCTGACAGGAGAA.............................................................................................................................................. |
29 |
2 |
1 |
1.00 |
1 |
1 |
| ..........................................................................TCGCACGAGAGCGCCATCTGACAGGAGT............................................................................................................................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................AATGATCGTAACTCCTTGCTGCACAAAGCCT.......................................................................................................................................................................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................TTCTCAATGATCGTAACTCCTTGCTGCAC................................................................................................................................................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................CTCGCACGAGAGCGCCATCTGACAGGAGGT.............................................................................................................................................. |
30 |
1 |
1 |
1.00 |
1 |
1 |