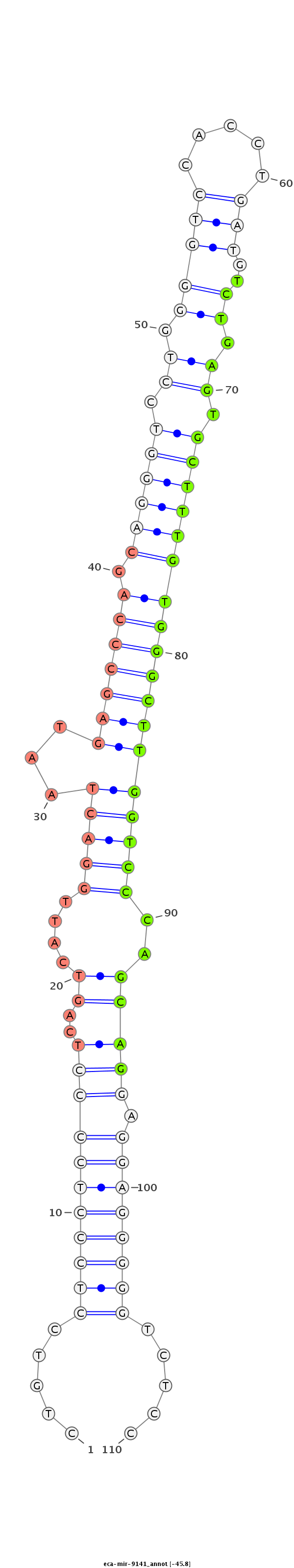
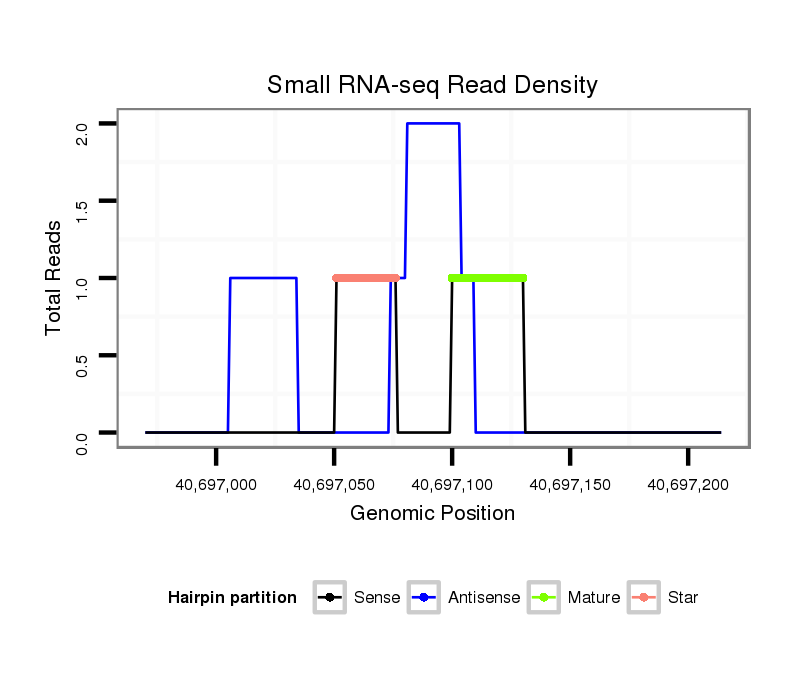
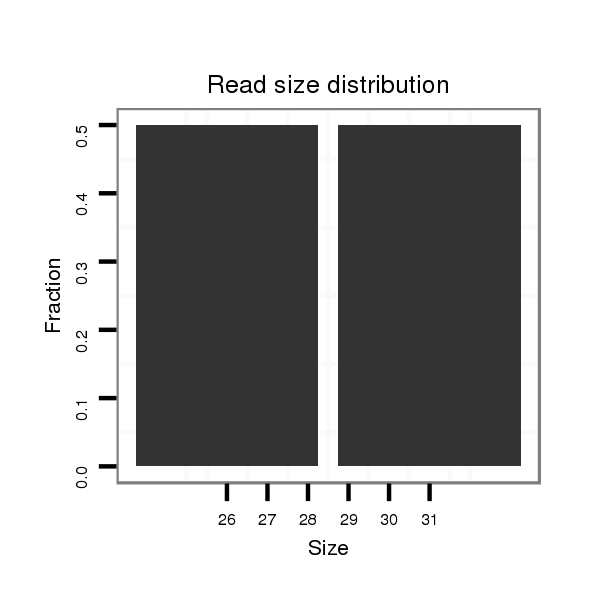
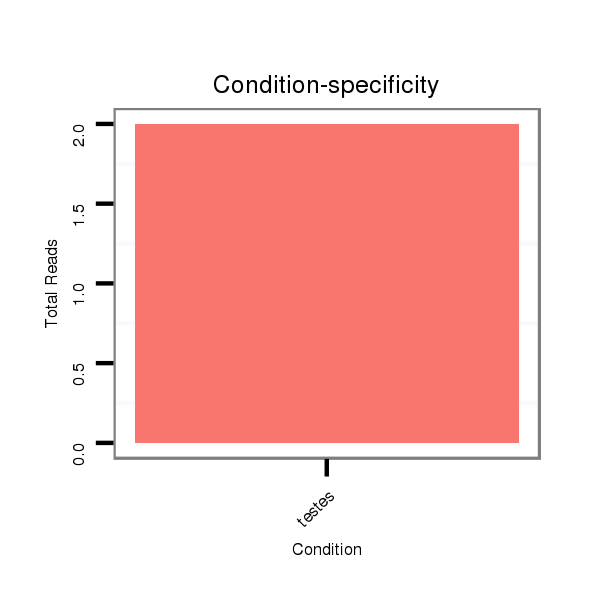
ID:eca-mir-9141 |
Coordinate:chr13:40697020-40697164 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
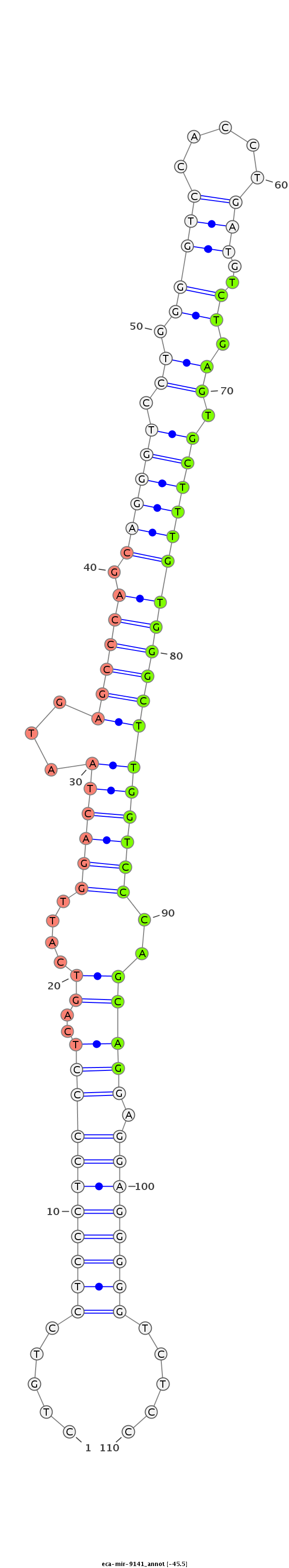
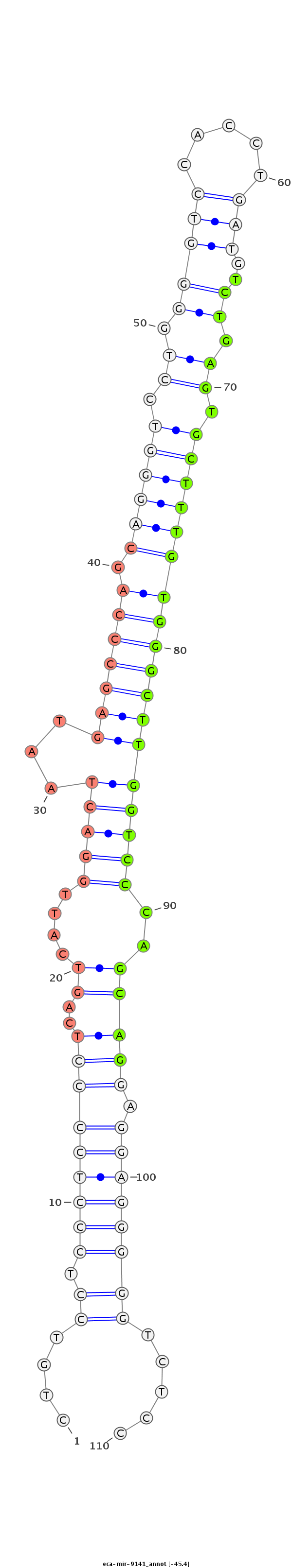
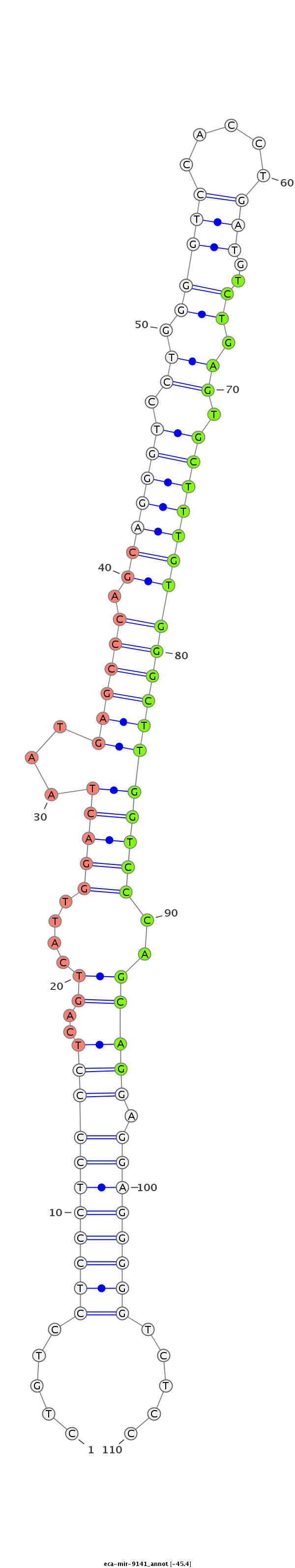
| -45.5 | -45.4 | -45.4 |
 |
 |
 |
|
CGCCACTGAGCTTCCACTGGGCCTGCGTGGCCTCAACACAGGTCAGCTGTGTGGCAGCACAGGGGGCTGTCCTCCCTCCCCTCAGTCATTGGACTAATGAGCCCAGCAGGGTCCTGGGGTCCACCTGATGTCTGAGTGCTTTGTGGGCTTGGTCCCAGCAGGAGGAGGGGGTCTCCCCAGCTGTCCCCTGGCCAGGCTCTGCTTTGAGGCCAGGCTGGCTTCTCACCACCCCCTTGTTGTGGGCT
******************************************************************.....(((((((((((..((....(((((...(((((((.((((((.((.(((((.....)))..)).)).))))))))))))))))))..))))).)))))))).....********************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ..................................................................................................................................TCTGAGTGCTTTGTGGGCTTGGTCCCAGCAG.................................................................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| .................................................................................TCAGTCATTGGACTAATGAGCCCAGC.......................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
|
GCGGTGACTCGAAGGTGACCCGGACGCACCGGAGTTGTGTCCAGTCGACACACCGTCGTGTCCCCCGACAGGAGGGAGGGGAGTCAGTAACCTGATTACTCGGGTCGTCCCAGGACCCCAGGTGGACTACAGACTCACGAAACACCCGAACCAGGGTCGTCCTCCTCCCCCAGAGGGGTCGACAGGGGACCGGTCCGAGACGAAACTCCGGTCCGACCGAAGAGTGGTGGGGGAACAACACCCGA
*********************************************************************.....(((((((((((..((....(((((...(((((((.((((((.((.(((((.....)))..)).)).))))))))))))))))))..))))).)))))))).....****************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ........................................................................................................TCGTCCCAGGACCCCAGGTGGACTACAGAC............................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................GTGTCCAGTCGACACACCGTCGTGTCCCC.................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................................AGGACCCCAGGTGGACTACAGACTCACGA......................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:40 AM