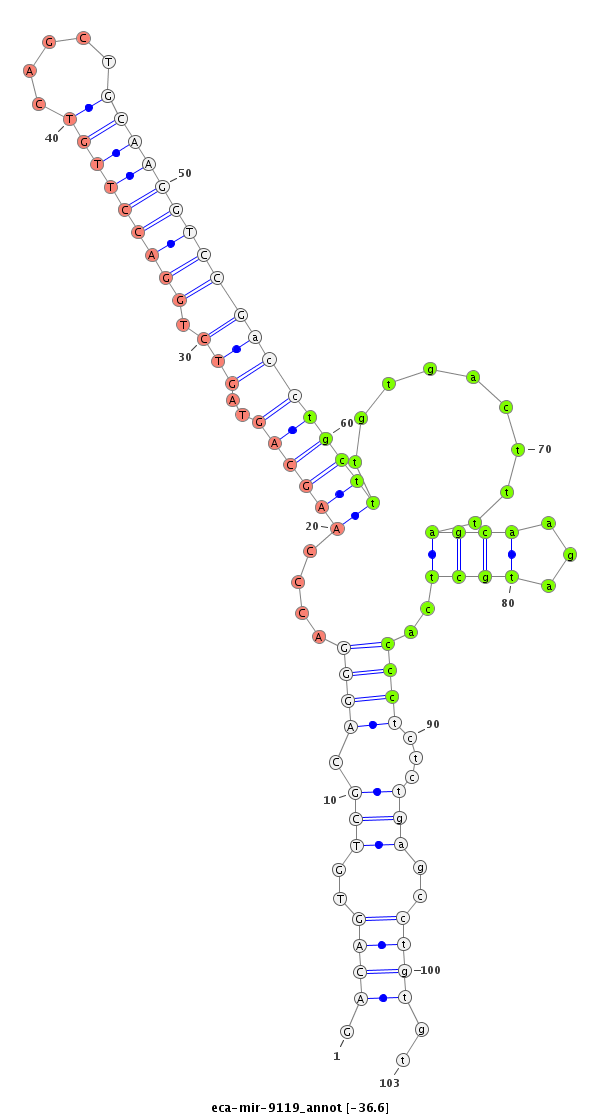
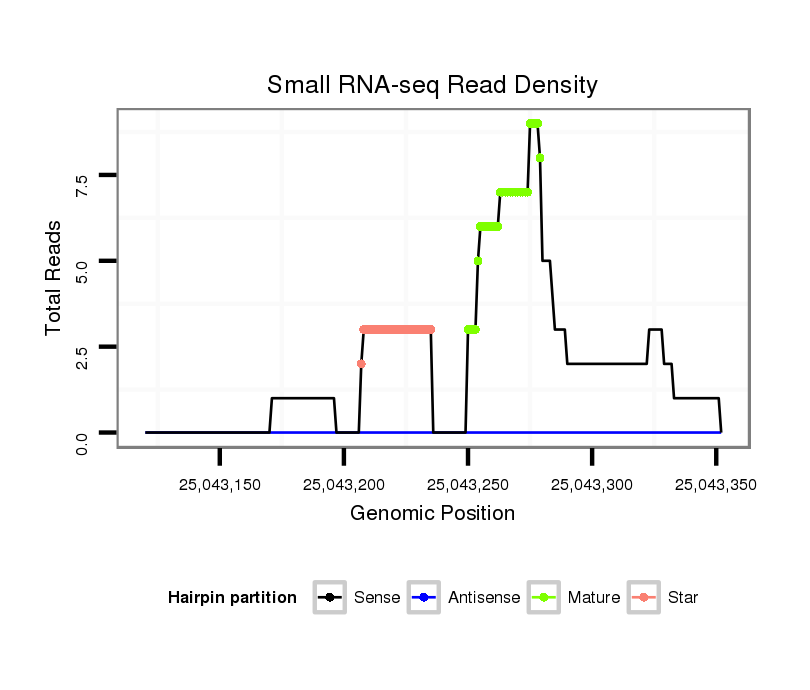
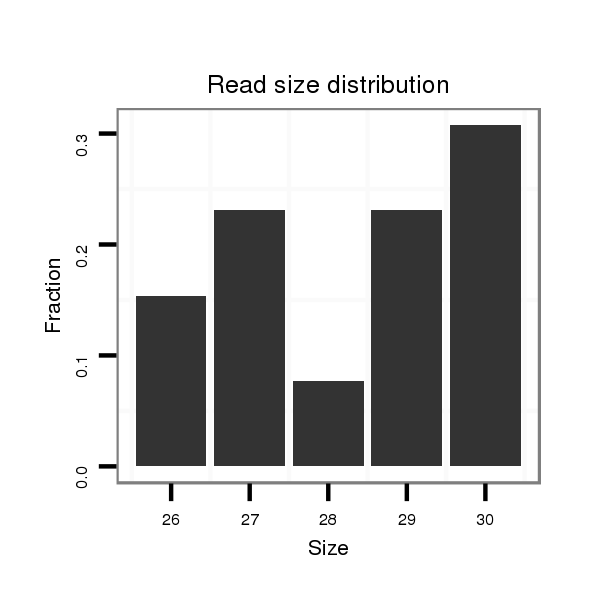
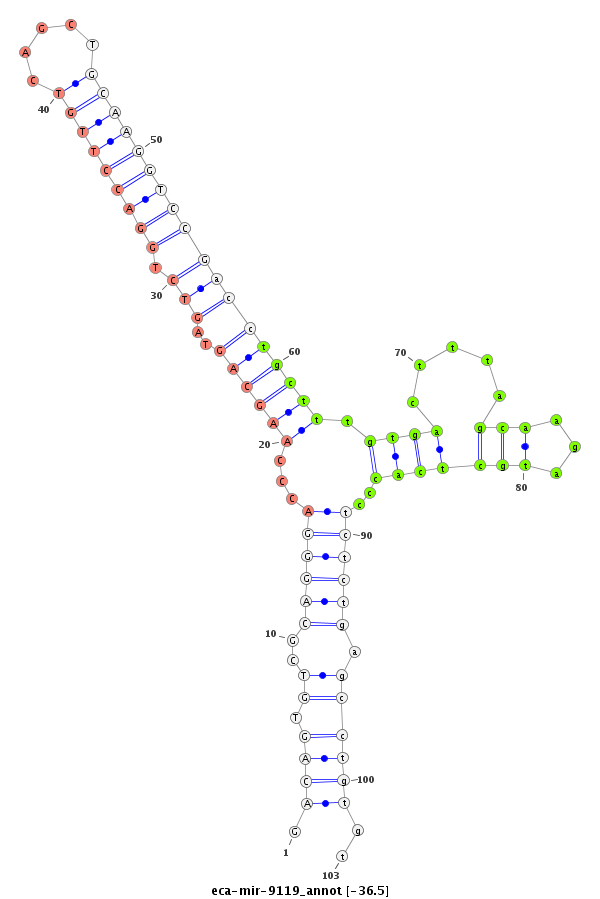
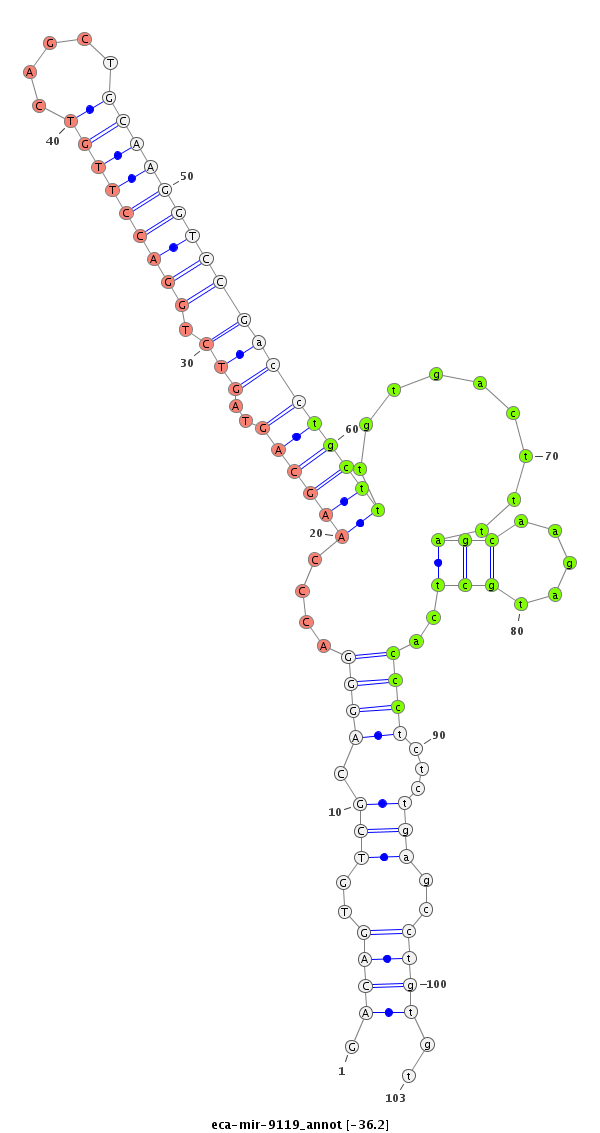
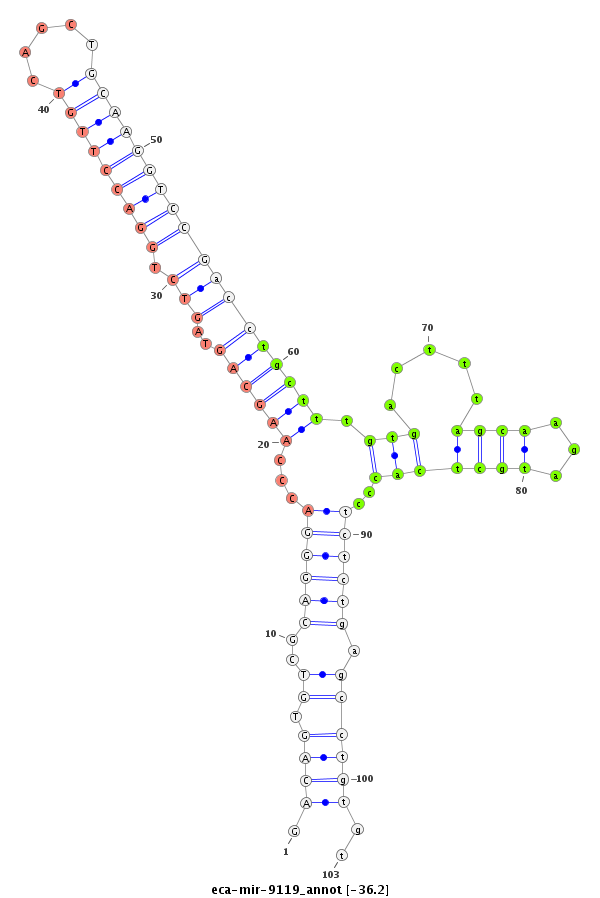
| ..................................................................................................................................TGCTTTGTGACTTTAGCAAGATGCTCACCC......................................................................... |
30 |
0 |
1 |
2.00 |
2 |
2 |
| .......................................................................................ACCCAAGCAGTAGTCTGGACCTTGTCAGC..................................................................................................................... |
29 |
0 |
1 |
2.00 |
2 |
2 |
| ...........................................................................................................................................................CACCCTCTCTGAGCCTGTGTCCTCATC................................................... |
27 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................................................................................................TTGTGACTTTAGCAAGATGCTCACCCTCTC..................................................................... |
30 |
0 |
1 |
1.00 |
1 |
1 |
| ........................................................................................CCCAAGCAGTAGTCTGGACCTTGTCAGC..................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................................................................................TCCCAAACAGGTTTGCCACAGTTGCCT........................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TGCTTTGTGACTTTAGCAAGATGCTCACC.......................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................................................TAGCAAGATGCTCACCCTCTCTGAGCC............................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................................................................................TCCCAAACAGGTTTGCCACAGTTGCCTTGCC.................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ............................................................................................................................CCGACCTGCTTTGTGACTTCAGCAAGA.................................................................................. |
27 |
1 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................................................................................................TTGCCTTGCCAGGTTGCCAAGATGATTCT. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................................................ACCCAAGCAGTAGTCTGGACCGTGTCAGC..................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
| ...................................................CACATGTGGGAAGGGGCGAGTGACAG............................................................................................................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................................................CCCCAAGCAGTAGTCTGGACCTTGTCAGCTG................................................................................................................... |
31 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................................TGTGACTTTAGCAAGATGCTCACCCTCTCT.................................................................... |
30 |
0 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................................................TTGTGACTTTAGCAAGATGCTCACCC......................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |