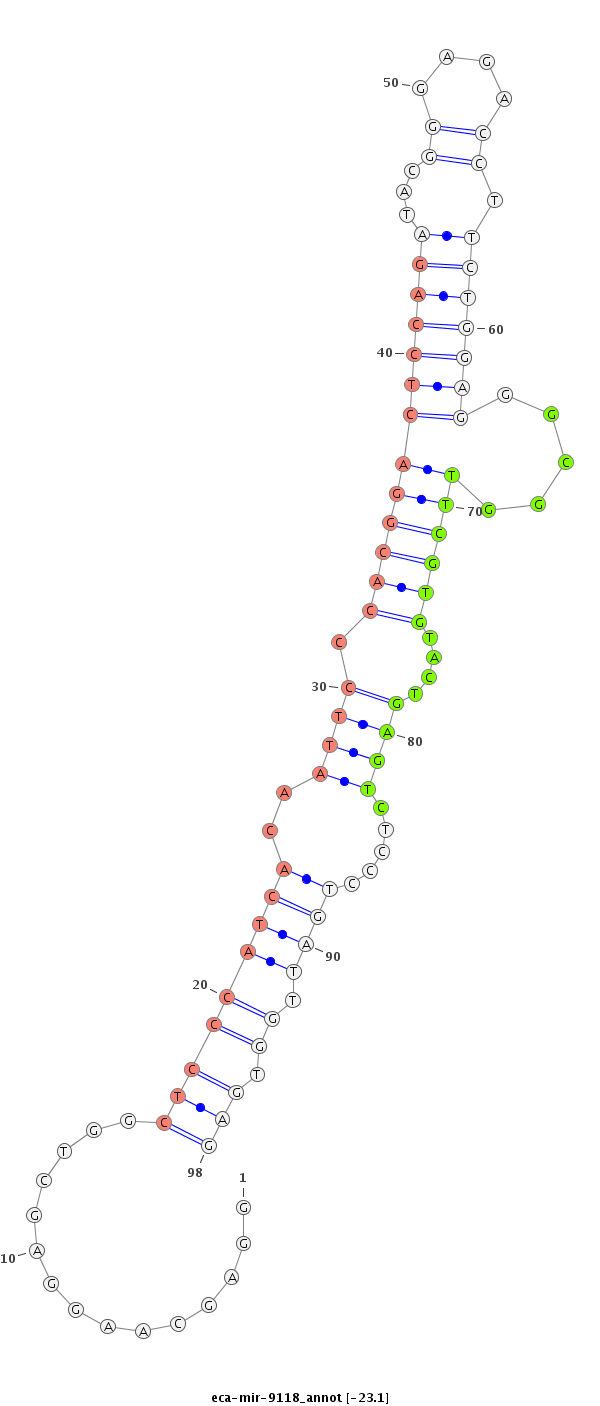
ID:eca-mir-9118 |
Coordinate:chr12:3391316-3391430 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
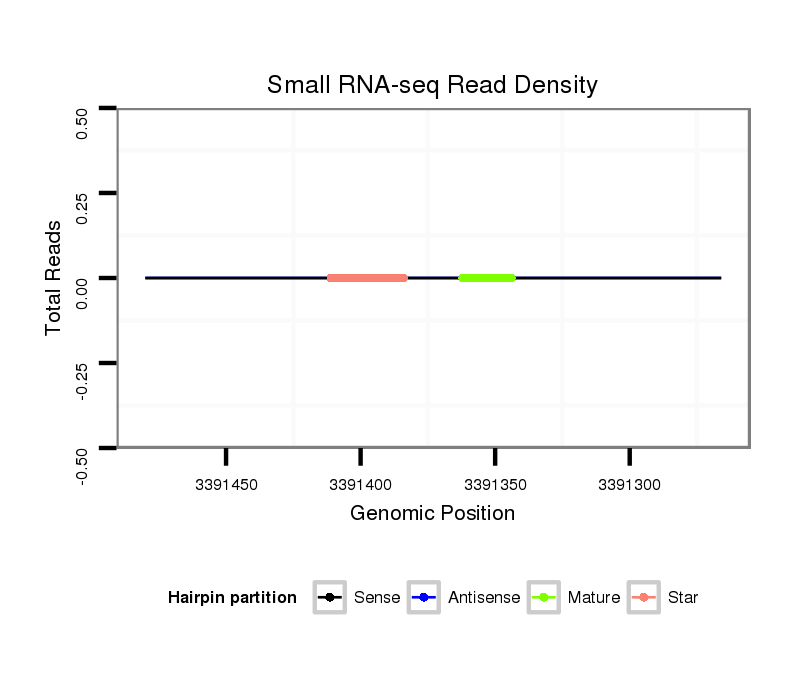
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
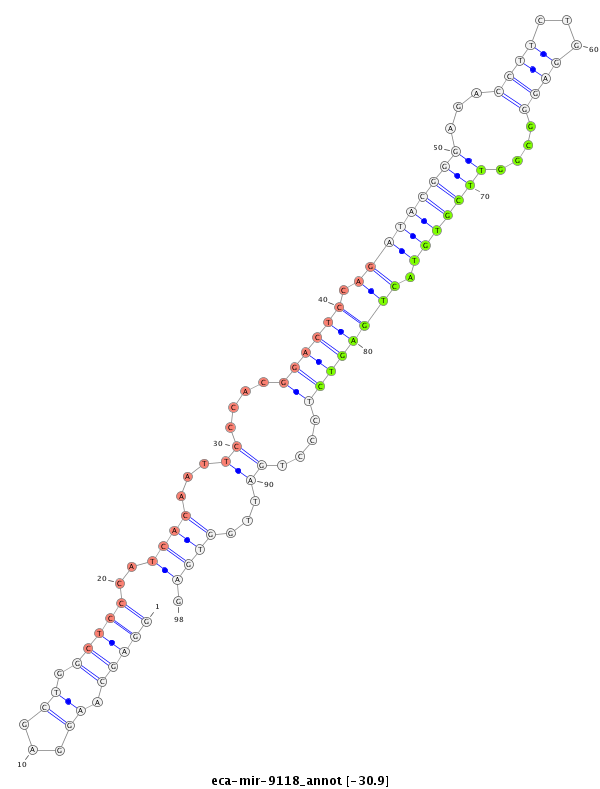
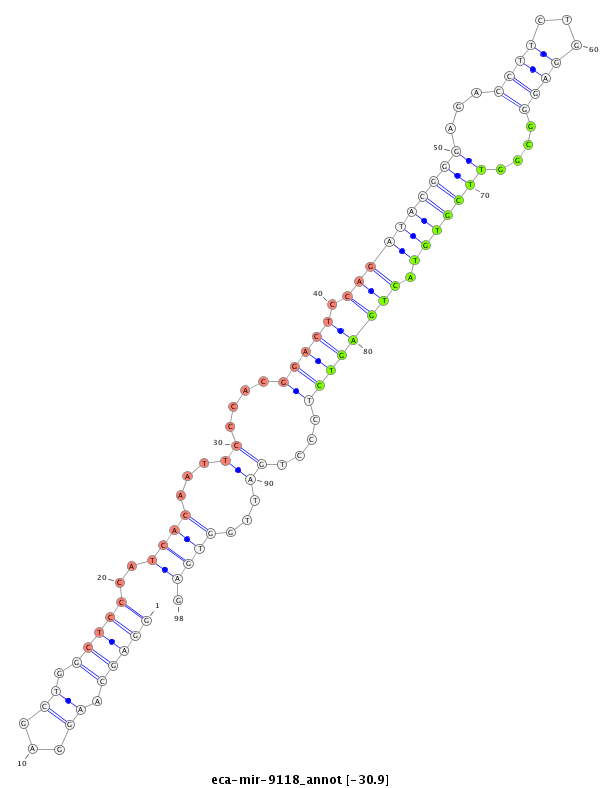
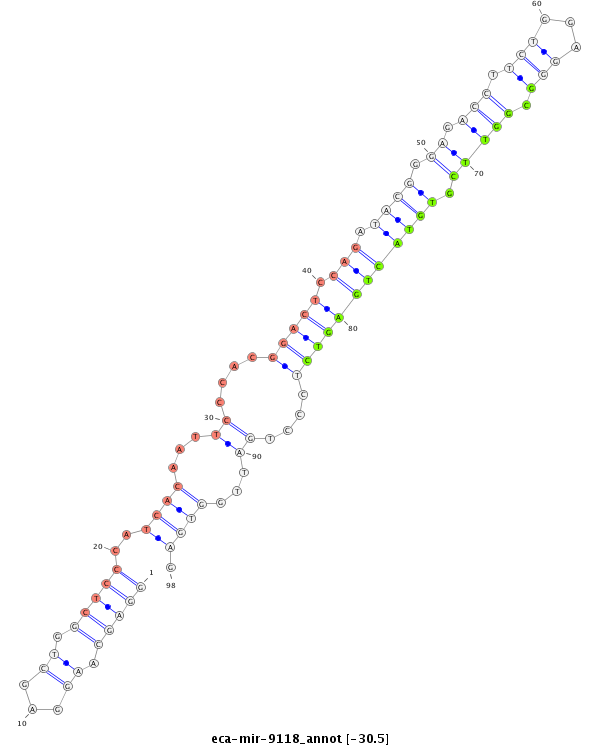
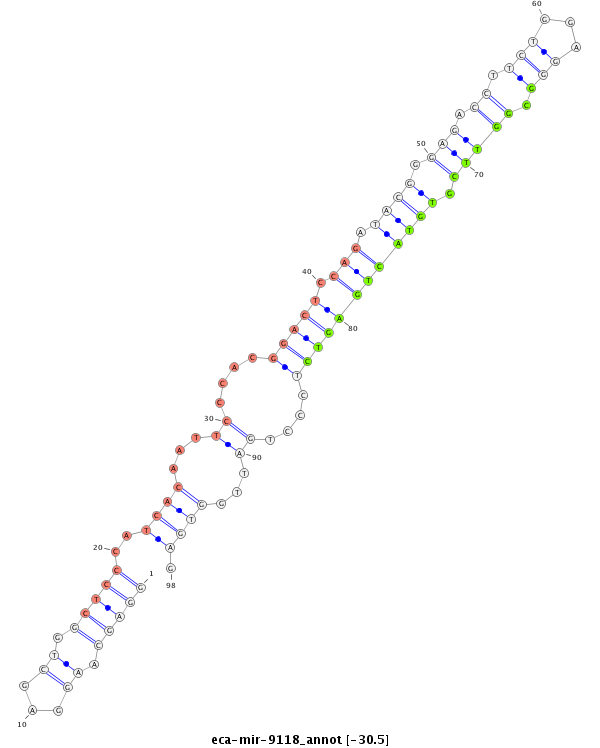
| -30.9 | -30.9 | -30.5 | -30.5 |
 |
 |
 |
 |
| mature | star |
|
GGCAATGTGGCTGGGCCACTCAGCACATAGCCTTCAGAGGCTGGGGTTAGGCTGGGAGCAAGGAGCTGGCTCCCATCACAATTCCCACGGACTCCAGATACGGGAGACCTTCTGGAGGGCGGTTCGTGTACTGAGTCTCCCTGATTGGTGAGCCATTTCCCTTCCGTGGGAGGAACATGGAAAGACAGTCACGTTTGCTTTCCCTGTGTGTGTTT
******************************************************...............(((((((((..((((.(((((((((((((...((....)).))))))).....))))))....)))).....)))).)).)))*************************************************************** |
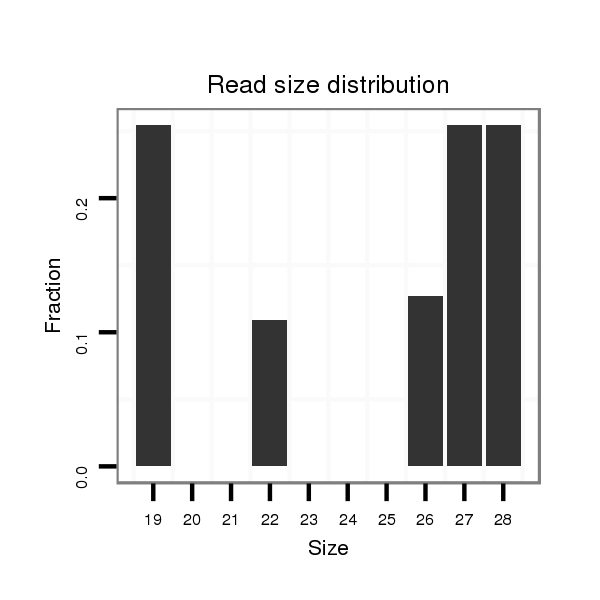
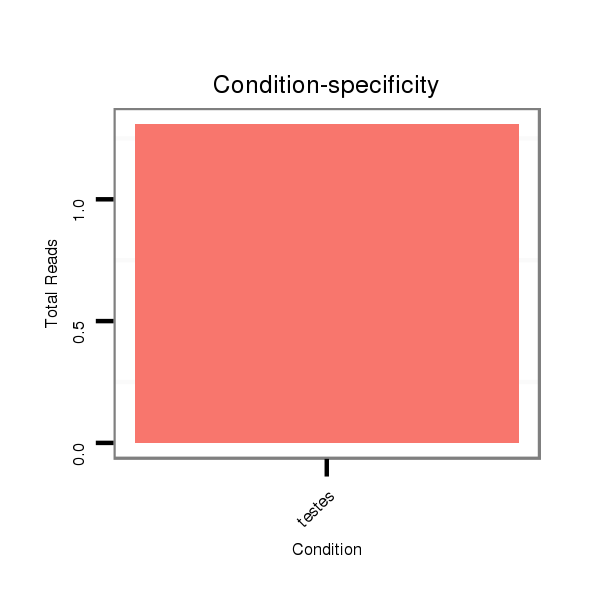
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ......................................................................................................................GCGGTTCGTGTACTGAGTC.............................................................................. | 19 | 0 | 3 | 0.33 | 1 | 1 |
| ............................................................................................................................CGTGTACTGAGTCTCCCTGATTGGTGA................................................................ | 27 | 0 | 6 | 0.33 | 2 | 2 |
| ..........................................................................................................................TTCGTGTACTGAGTCTCCCTGATTGGTGAGCCT............................................................ | 33 | 1 | 5 | 0.20 | 1 | 1 |
| ...........................................................................TCACAATTCCCACGGACTCCAGATACGGGAA............................................................................................................. | 31 | 1 | 5 | 0.20 | 1 | 1 |
| ...........................................................................................................................TCGTGTACTGAGTCTCCCTGATTGGTGT................................................................ | 28 | 1 | 6 | 0.17 | 1 | 1 |
| .....................................................................CTCCCATCACAATTCCCACGGACTCCAG...................................................................................................................... | 28 | 0 | 6 | 0.17 | 1 | 1 |
| ...........................................................................................................................TCGTGTACTGAGTCTCCCTGATTGGTGA................................................................ | 28 | 0 | 6 | 0.17 | 1 | 1 |
| ...........................................................................................................................TCGTGTACTGAGTCTCCCTGATTGGT.................................................................. | 26 | 0 | 6 | 0.17 | 1 | 1 |
| ...........................................................................TCACAATTCCCACGGACTCCAGATC................................................................................................................... | 25 | 1 | 6 | 0.17 | 1 | 1 |
| ..............................................................................................................................TGTACTGAGTCTCCCTGATTGG................................................................... | 22 | 0 | 7 | 0.14 | 1 | 1 |
|
CCGTTACACCGACCCGGTGAGTCGTGTATCGGAAGTCTCCGACCCCAATCCGACCCTCGTTCCTCGACCGAGGGTAGTGTTAAGGGTGCCTGAGGTCTATGCCCTCTGGAAGACCTCCCGCCAAGCACATGACTCAGAGGGACTAACCACTCGGTAAAGGGAAGGCACCCTCCTTGTACCTTTCTGTCAGTGCAAACGAAAGGGACACACACAAA
***************************************************************...............(((((((((..((((.(((((((((((((...((....)).))))))).....))))))....)))).....)))).)).)))****************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .........................................................................................................................CAAGCACATGACTCAGAGGGACTAACCACT................................................................ | 30 | 0 | 6 | 0.33 | 2 | 2 |
| ...........................................................................................................................AGCACATGACTCAGAGGGACTAACCACT................................................................ | 28 | 0 | 6 | 0.33 | 2 | 2 |
| ..........................................................................................................................................................................TCCTTGTACCTTTCTGTCAGTGCA..................... | 24 | 0 | 7 | 0.29 | 2 | 2 |
| .............................................................................................................................................................AGGGAAGGCACCCTCCTTGTACCTTTCTGT............................ | 30 | 0 | 4 | 0.25 | 1 | 1 |
| .........................................................................................................................CAAGCACGTGACTCAGAGGGACTAACCACT................................................................ | 30 | 1 | 6 | 0.17 | 1 | 1 |
| ..........................................................................TAGTGTTAAGGGTGCCTGAGGTCTAT................................................................................................................... | 26 | 0 | 6 | 0.17 | 1 | 1 |
| ........................................................................................................................CCAAGCACATGACTCAGAGGGACTAACCAC................................................................. | 30 | 0 | 6 | 0.17 | 1 | 1 |
| .........................................................................................................................AAAGCACATGACTCAGAGGGACTAACC................................................................... | 27 | 1 | 6 | 0.17 | 1 | 1 |
| ..........................................................................................................................................................................TCCTTGTACCTTTCTGTCAGTGCAAAC.................. | 27 | 0 | 7 | 0.14 | 1 | 1 |
| .................................AGTCTCCGACCCCAAGCCGACCCTCGTT.......................................................................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 11:39 AM