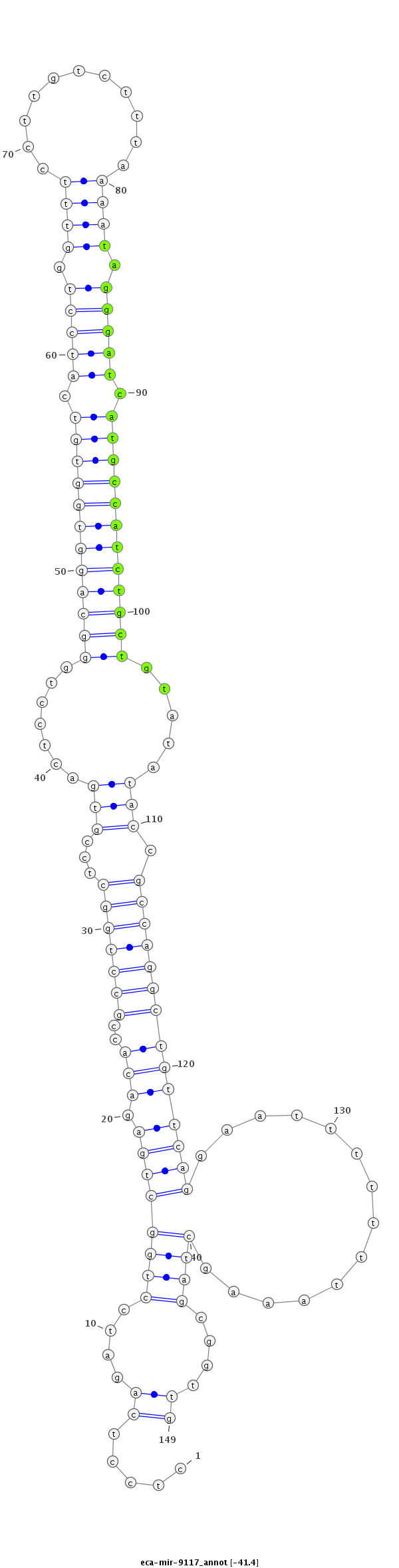
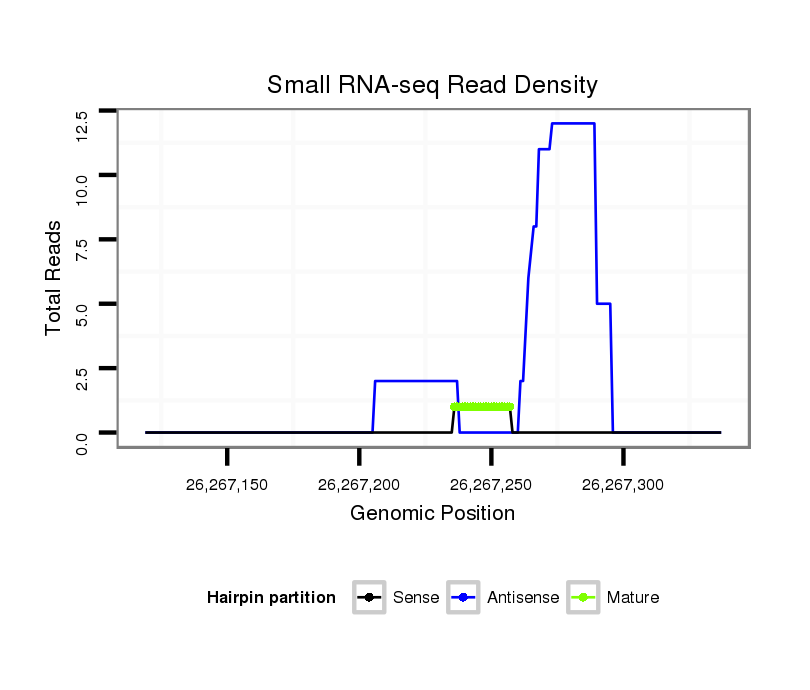
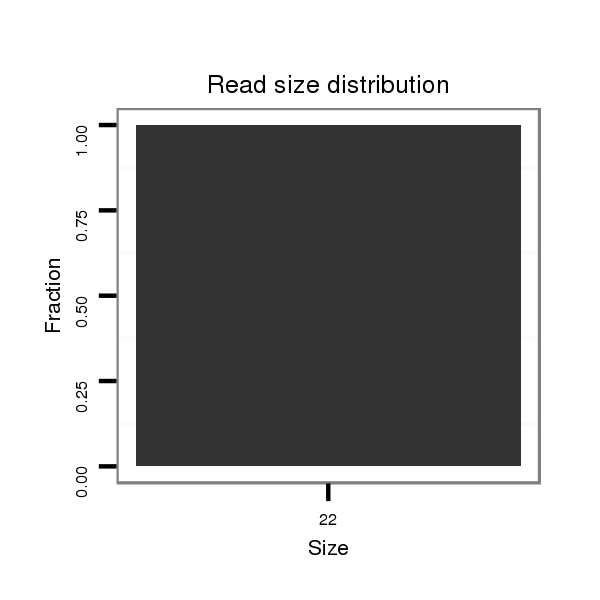
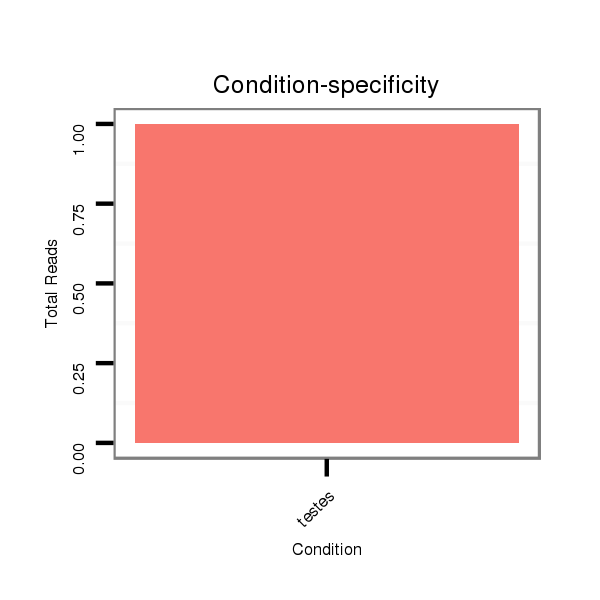
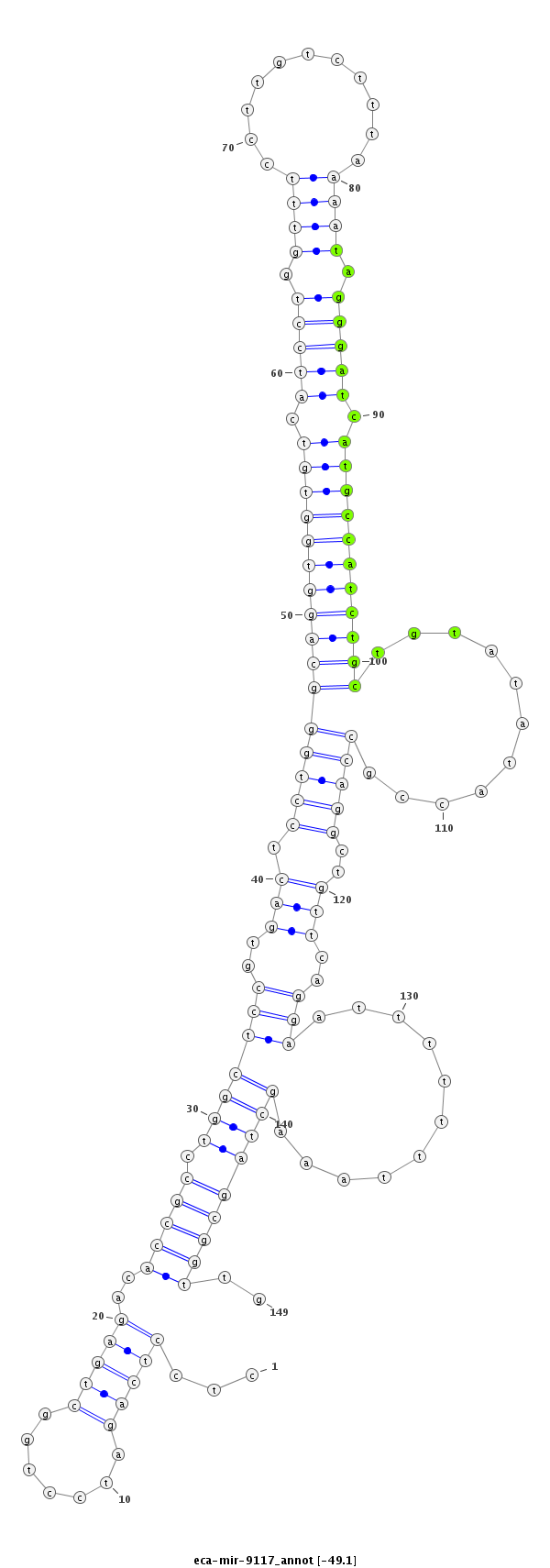
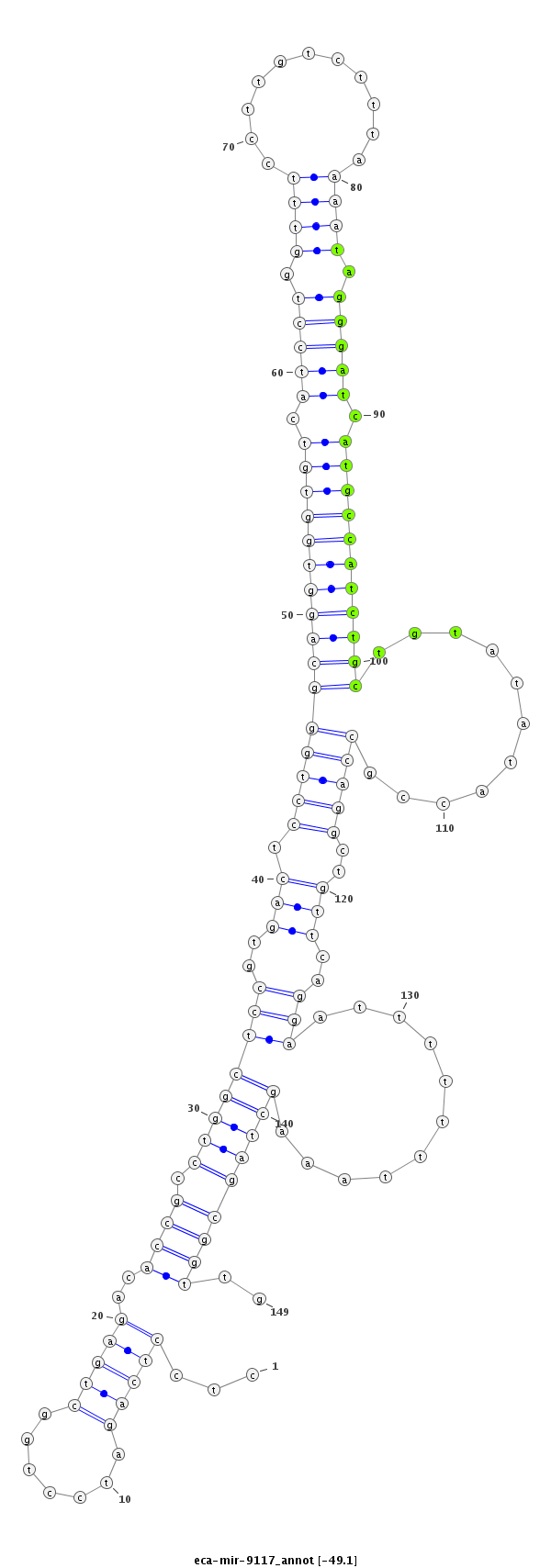
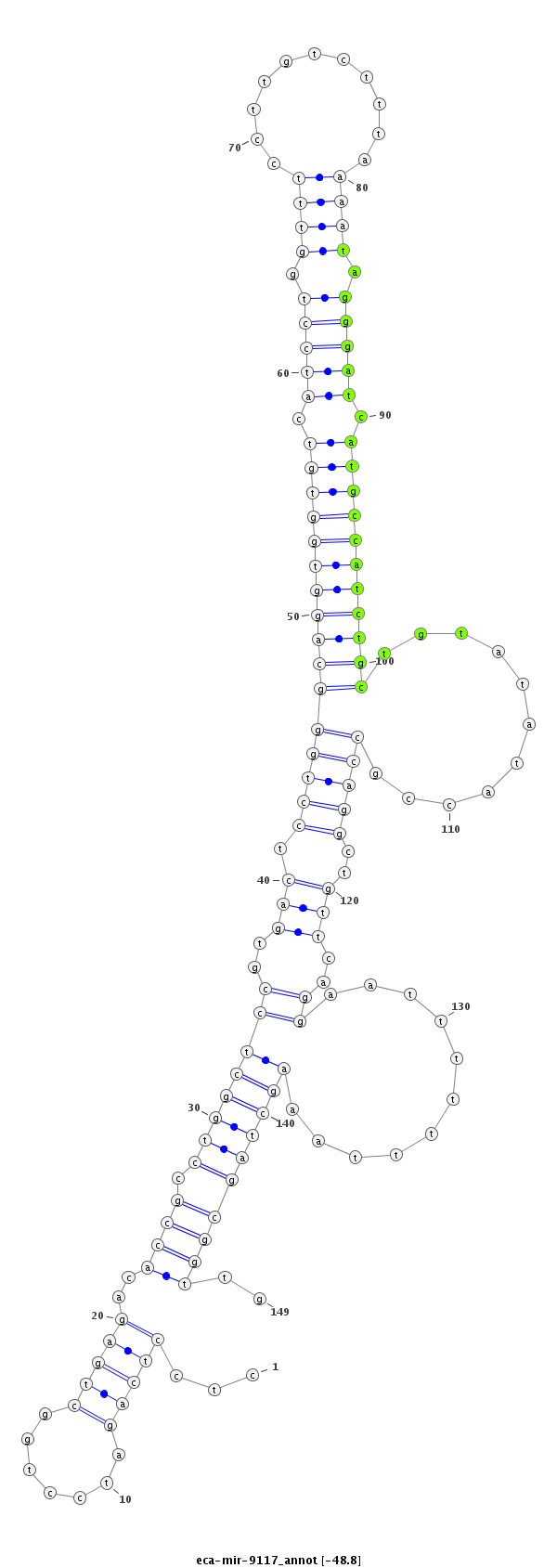
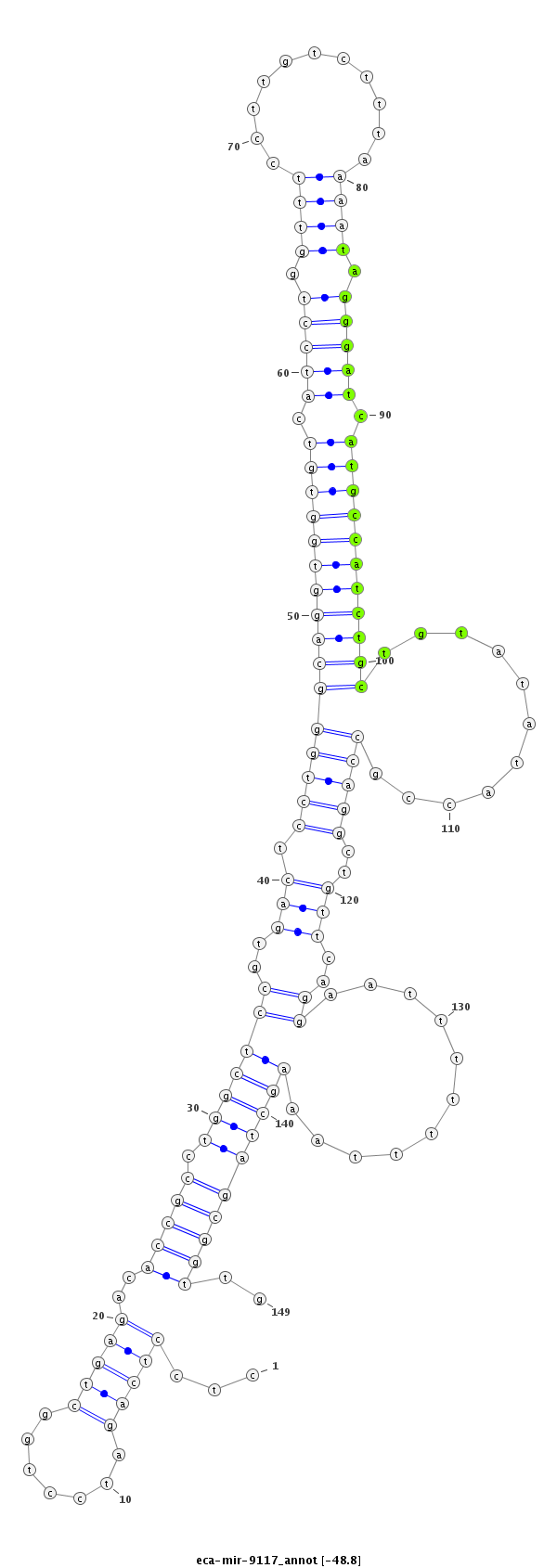
ID:eca-mir-9117 |
Coordinate:chr12:26267169-26267287 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -49.1 | -49.1 | -48.8 | -48.8 |
 |
 |
 |
 |
|
agaaggagttaggctccgggggaaagagtcttggcctcctcagatcctggctgagacaccgcctggctccgtgactcctgggcaggtggtgtcatcctggtttccttgtctttaaaatagggatcatgccatctgctgtatataccgccaggctgttcaggaatttttttaaagctagcggttgtgaaaacacctGTCACTTAGGCATTTGCTAATCAA
***********************************.....((....((((((((.(((..(((((((...(((.......((((((((((((.(((((.((((...........)))).))))).)))))))))))).....))).))))))))))))))..............))))....))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .....................................................................................................................TAGGGATCATGCCATCTGCTGT................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 |
|
TgTTggTgttTggGtGGgggggTTTgTgtGttggGGtGGtGTgTtGGtggGtgTgTGTGGgGGtggGtGGgtgTGtGGtgggGTggtggtgtGTtGGtggtttGGttgtGtttTTTTtTgggTtGTtgGGTtGtgGtgtTtTtTGGgGGTggGtgttGTggTTtttttttTTTgGtTgGggttgtgTTTTGTGGtCAGTGAATCCGTAAACGATTAGTT
***********************************.....((....((((((((.(((..(((((((...(((.......((((((((((((.(((((.((((...........)))).))))).)))))))))))).....))).))))))))))))))..............))))....))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .....................................................................................................................................................TCCGACAAGTCCTTAAAAAAATTTCGAT.......................................... | 28 | 0 | 1 | 3.00 | 3 | 3 |
| .................................................................................................................................................GCGGTCCGACAAGTCCTTAAAAAAAT................................................ | 26 | 0 | 1 | 2.00 | 2 | 2 |
| ................................................................................................................................................GGCGGTCCGACAAGTCCTTAAAAAAAT................................................ | 27 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................CCACAGTAGGACCAAAGGAACAGAAATTTTAT.................................................................................................... | 32 | 0 | 1 | 2.00 | 2 | 2 |
| ..............................................................................................................................................ATGGCGGTCCGACAAGTCCTTAAAAAAAT................................................ | 29 | 0 | 1 | 2.00 | 2 | 2 |
| ...................................................................................................................................................GGTCCGACAAGTCCTTAAAAAAATTTCGAT.......................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................................CGGTCCGACAAGTCCTTAAAAAAAT................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................................................CAAGTCCTTAAAAAAATTTCGAT.......................................... | 23 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................................................................................................................TCCGACAAGTCCTTAAAAAATTTTCGAT.......................................... | 28 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................................CGGTCCGACAAGTCCT......................................................... | 16 | 0 | 5 | 0.20 | 1 | 1 |
Generated: 09/01/2015 at 11:38 AM