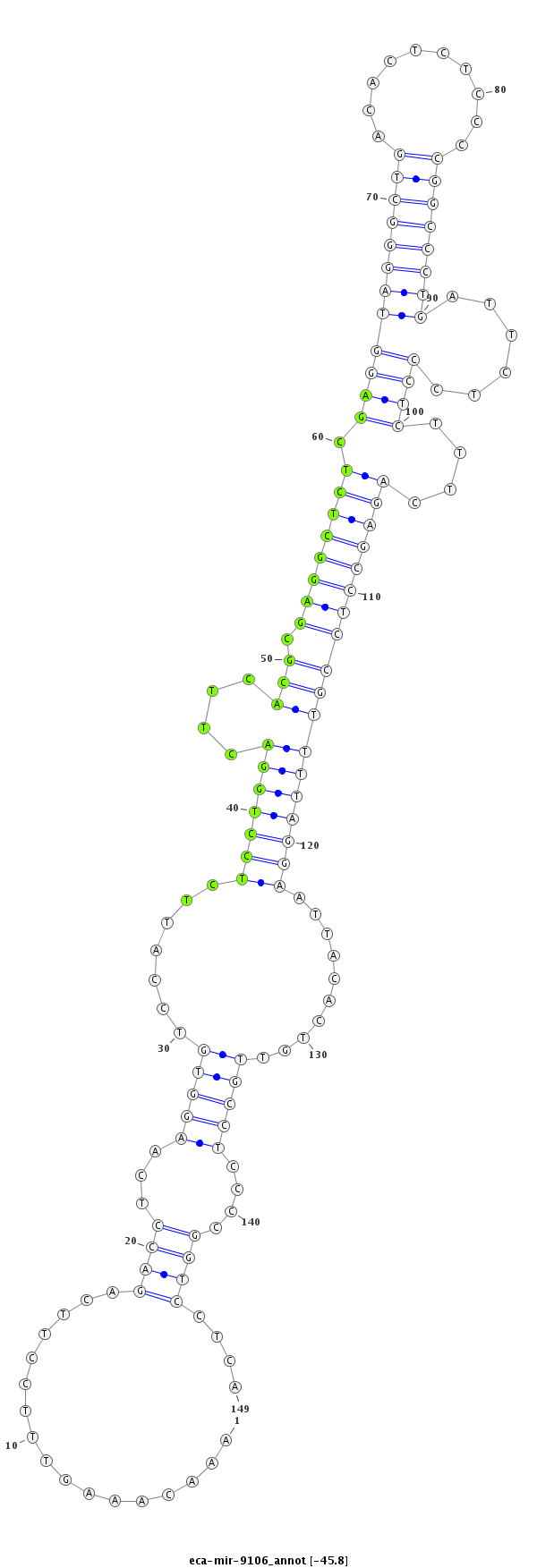
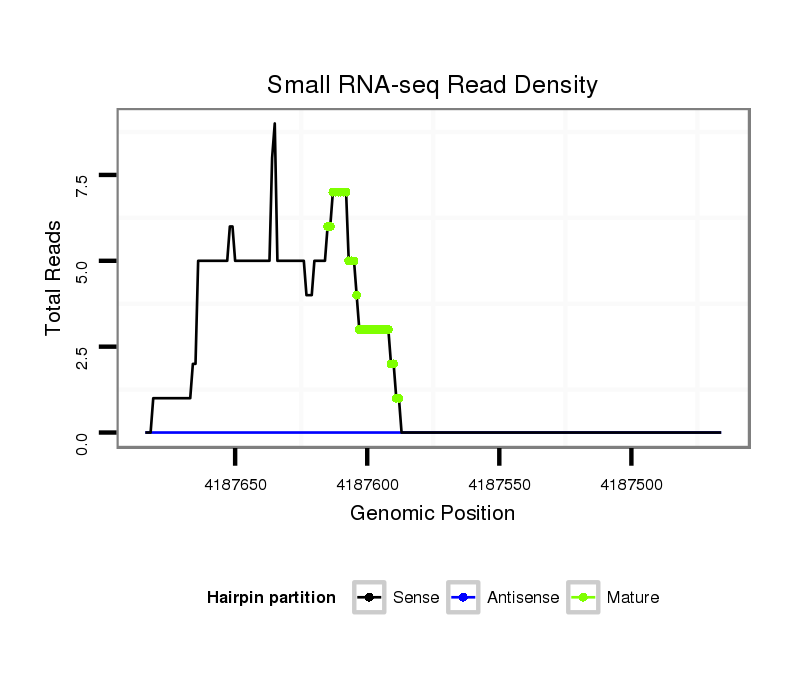
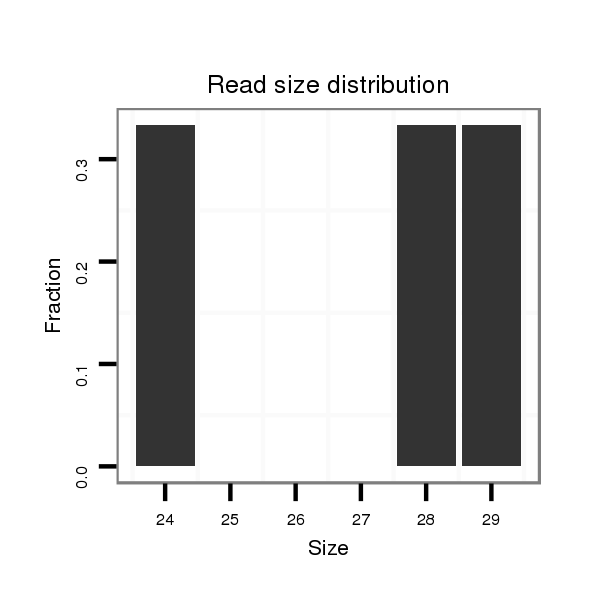
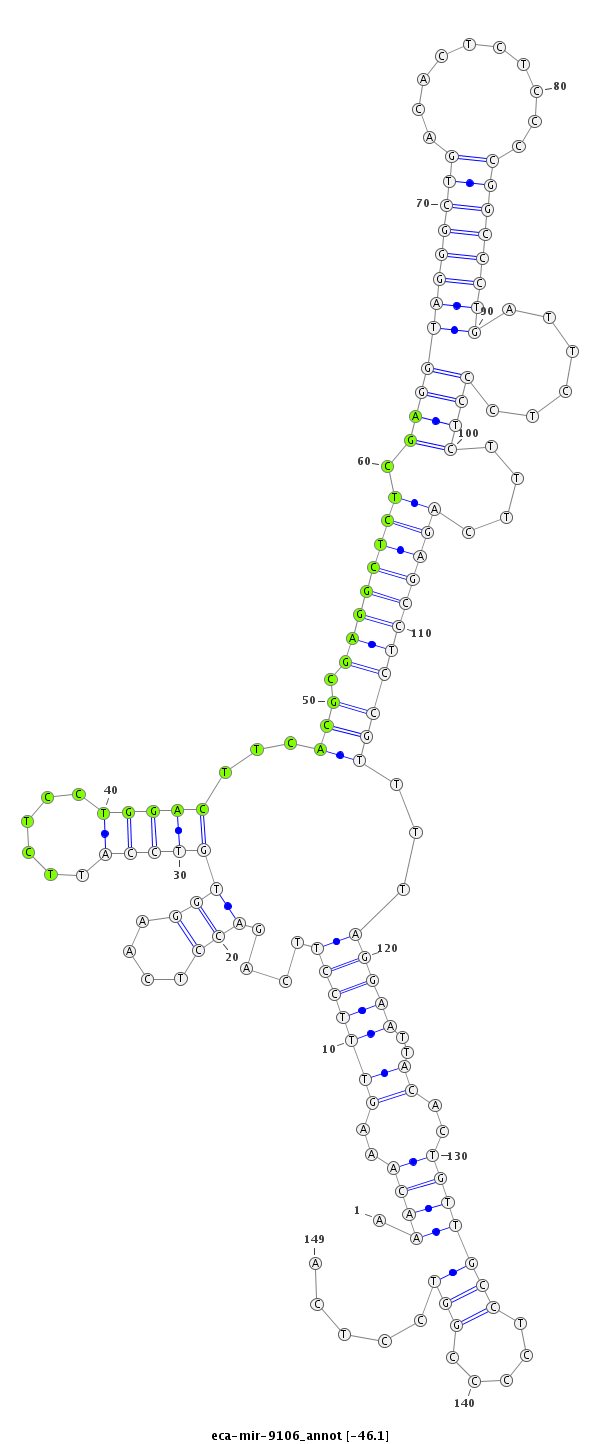
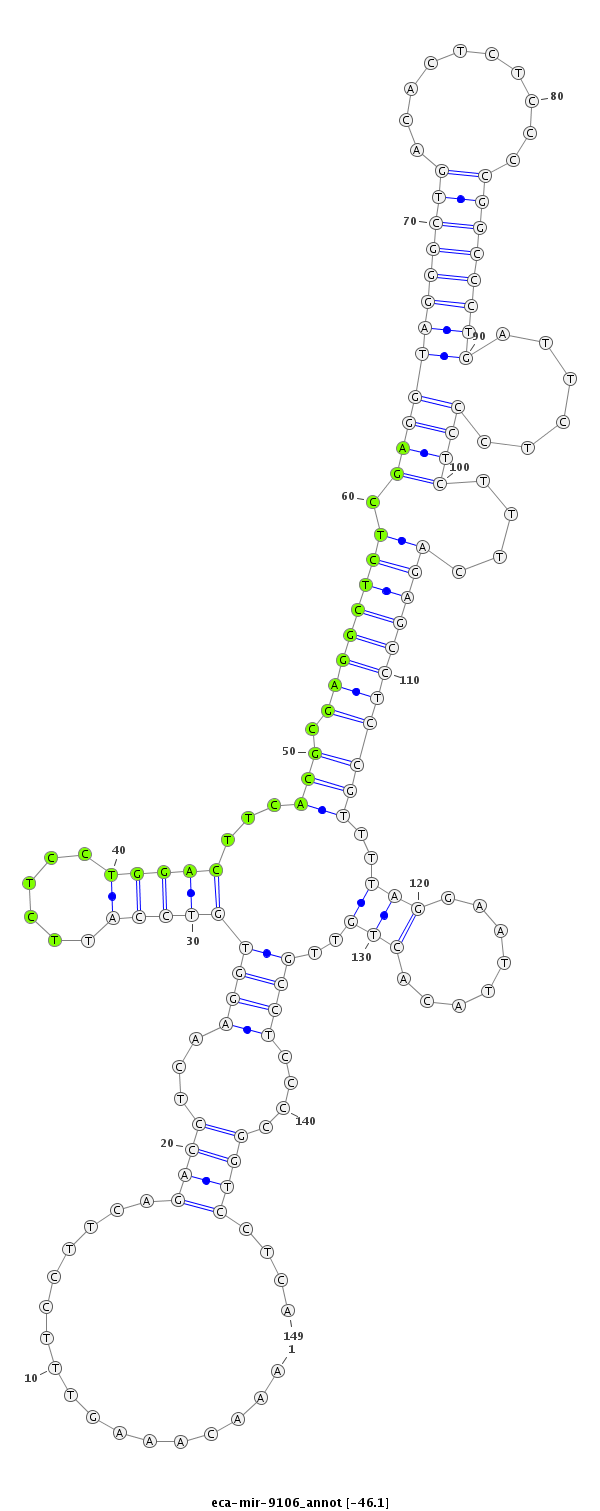
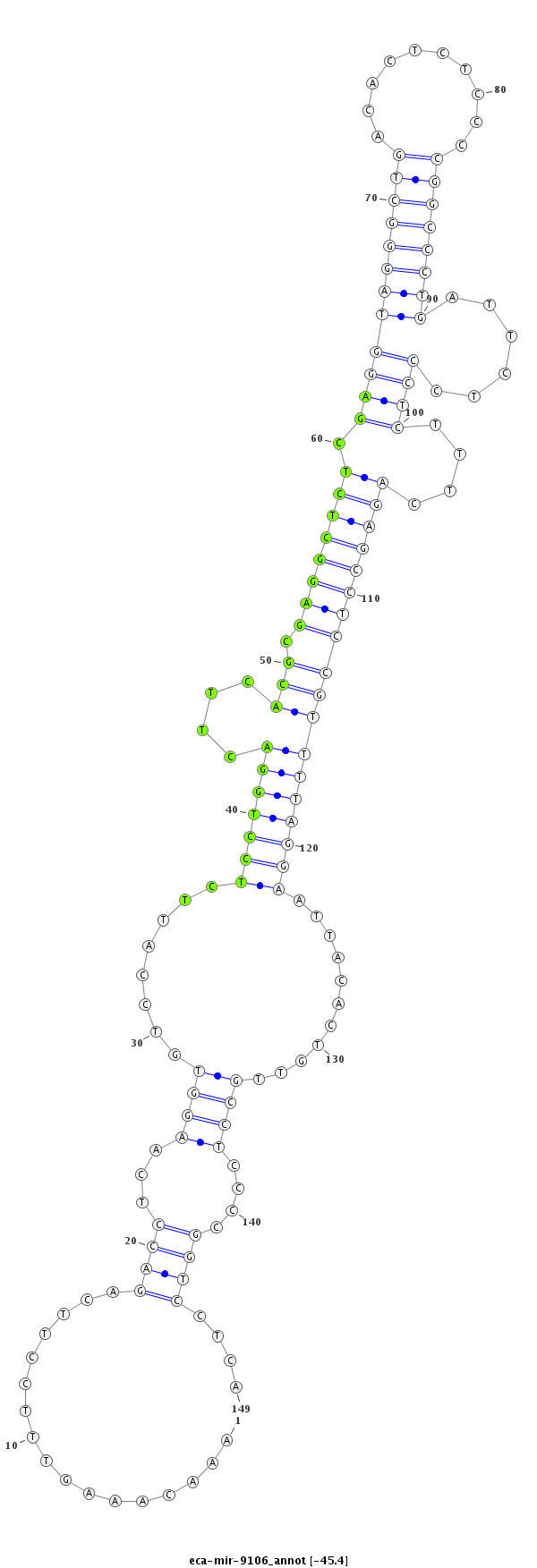
| ....................TAGAAGAGTCACTCCAAACAAAGTTTCCTT......................................................................................................................................................................... |
30 |
0 |
1 |
3.00 |
3 |
3 |
| ................................................TTCAGACCTCAAGGTGTCCATTCTCCTGG.............................................................................................................................................. |
29 |
0 |
1 |
2.00 |
2 |
2 |
| .................................................TCAGACCTCAAGGTGTCCATTCTCCTGGACT........................................................................................................................................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................TTCTCCTGGACTTCACGCGAGGCTCTCGATT........................................................................................................................ |
31 |
2 |
1 |
1.00 |
1 |
1 |
| ....................................................................TTCTCCTGGACTTCACGCGAGGCTCA............................................................................................................................. |
26 |
1 |
1 |
1.00 |
1 |
1 |
| ................................TCCAAACAAAGTTTCCTTCAGACCTCAAG.............................................................................................................................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................................TCCTGGACTTCACGCGAGGCTGTCGA.......................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
| .....................................................................TCTCCTGGACTTCACGCGAGGCTCTCGA.......................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................TCTCCTGGACTTCACGCGAGGCTCTCGTA......................................................................................................................... |
29 |
2 |
1 |
1.00 |
1 |
1 |
| ....................TAGAAGAGTGACTCCAAACAAAGTT.............................................................................................................................................................................. |
25 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................................................TTCTCCTGGACTTCACGCGAGGCTCTCT........................................................................................................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................................TCCTGGACTTCACGCGAGGCTCTC............................................................................................................................ |
24 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................TTCAGACCTCAAGGTGTCCATTCTCCTGGACTT.......................................................................................................................................... |
33 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................................TCACGCGAGGCTCTCGAGGTAGAAAT................................................................................................................. |
26 |
3 |
1 |
1.00 |
1 |
1 |
| ...TTGCCTGGAAAGCACTCTAGAAGAGTCACTC......................................................................................................................................................................................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................TCCATTCTCCTGGACTTCACGCGAGGCTC.............................................................................................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................TTCTCCTGGACTTCACGCGAGGCTCTCGAGT........................................................................................................................ |
31 |
1 |
1 |
1.00 |
1 |
1 |
| ..................TCTAGAAGAGTCACTCCAAACAAAGTTTCCTT......................................................................................................................................................................... |
32 |
0 |
1 |
1.00 |
1 |
1 |