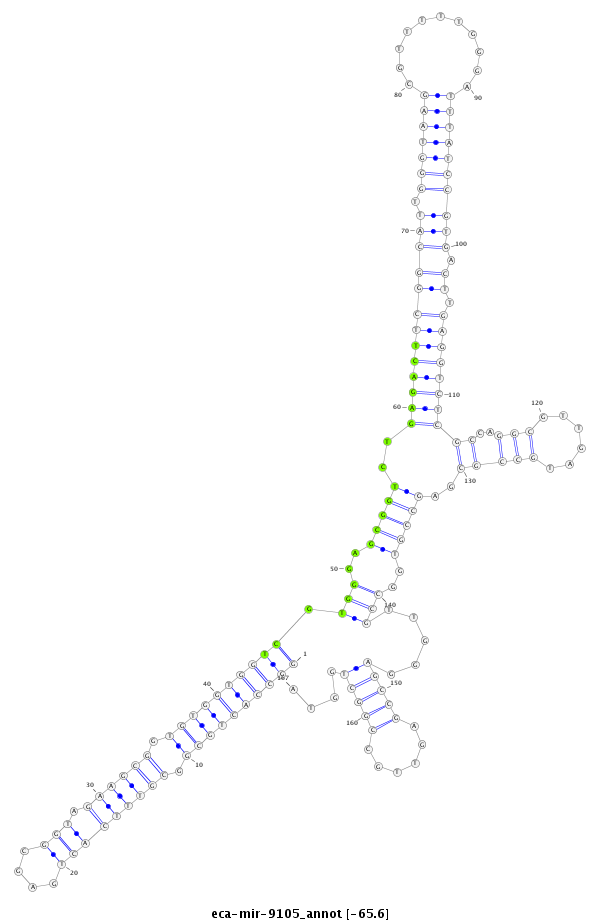
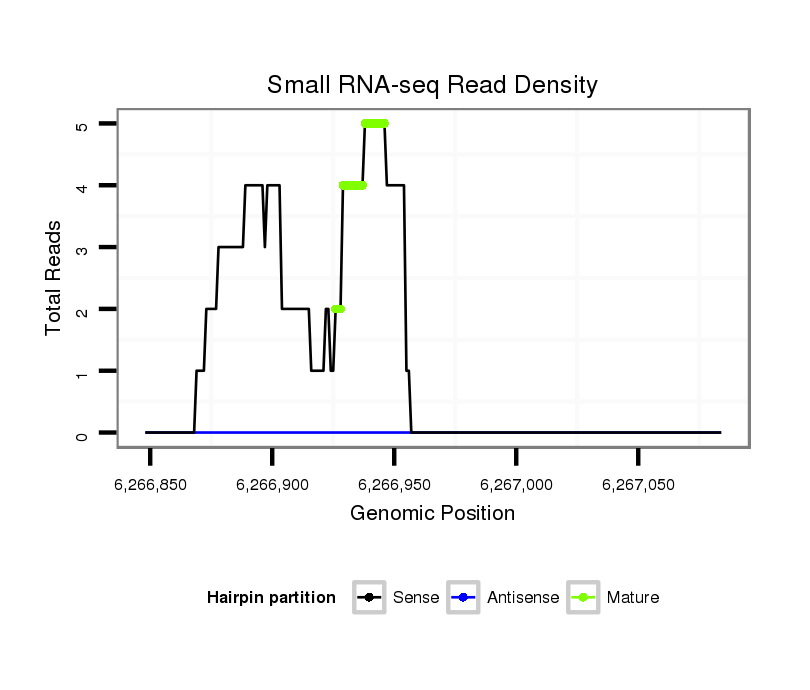
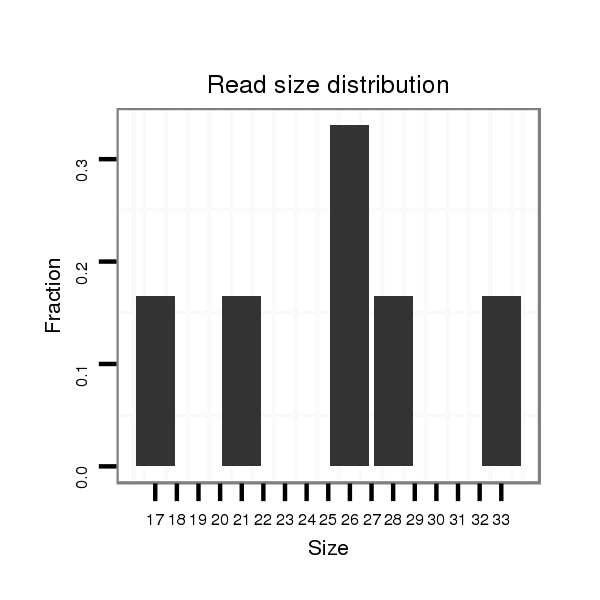
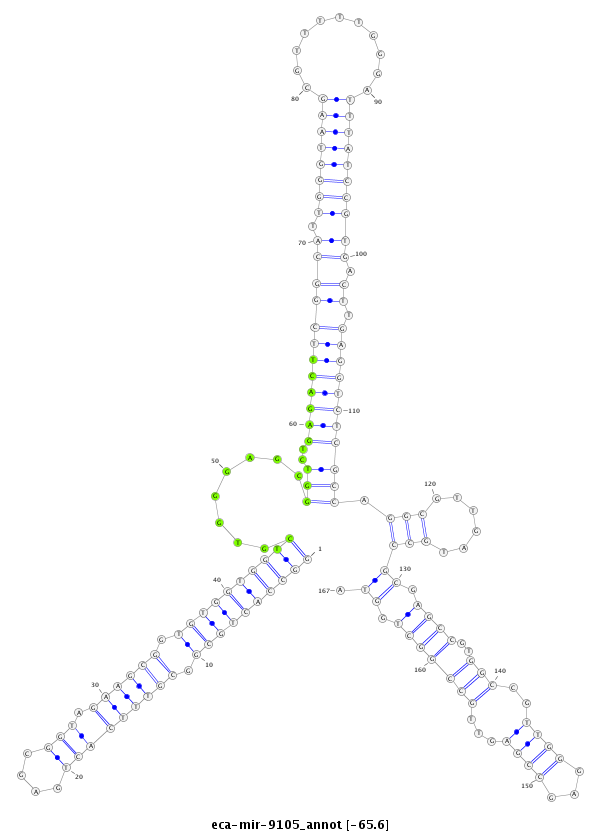
| ..............................................................................TCGTGGGAGCGGTCTGAGACT.......................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................CGTGGGAGCGGTCTGAGACGTCG....................................................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ........................................................................TGGTGGTCGTGGGAGCGGTCTGAGACTTCGGCATTGGGTG............................................................................................................................. |
40 |
1 |
1 |
1.00 |
1 |
1 |
| ................................CTTGGCCACTGCGGCGTTTCACTGAGCA................................................................................................................................................................................. |
28 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................TGGGAGCGGTCTGAGACTTCGGCATTGG................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................................AGCGGTCTGAGCCTTCGGCAT................................................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| ..........................................................................GTGGTCGTGGGAGCGGTCTGAGACTTCGGCATT.................................................................................................................................. |
33 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................TGCGGCGTTTCACTGAGCGGTAGAAGC......................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................TCACTGAGCGGTAGAAGCGGTGTGGT................................................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................TTCTTGGCCACTGCGGCGTTTCACTG..................................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
| .....................TGTTTTTGTTTCTTGGCCACTGCGGCGT............................................................................................................................................................................................ |
28 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................TCTGAGACTTCGGCATT.................................................................................................................................. |
17 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................TCACTGAGCGGTAGAAGCGGTGTGGTGGTT............................................................................................................................................................. |
30 |
1 |
1 |
1.00 |
1 |
1 |
| .........................TTTGTTTCTTGGCCACTGCGGCGTTTCACTG..................................................................................................................................................................................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................................TGGGAGCGGTCTGAGACTTCGGCATT.................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................TAACTGAGCGGTAGAAG.......................................................................................................................................................................... |
17 |
1 |
2 |
0.50 |
1 |
1 |