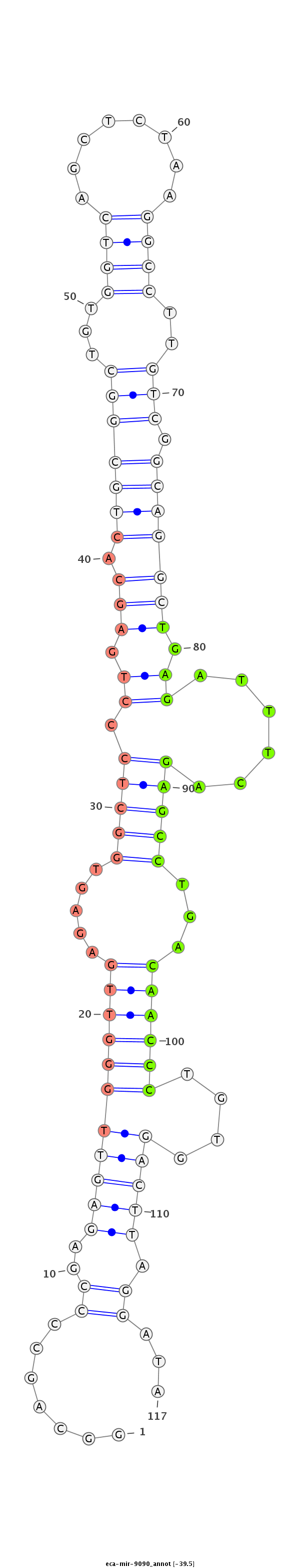
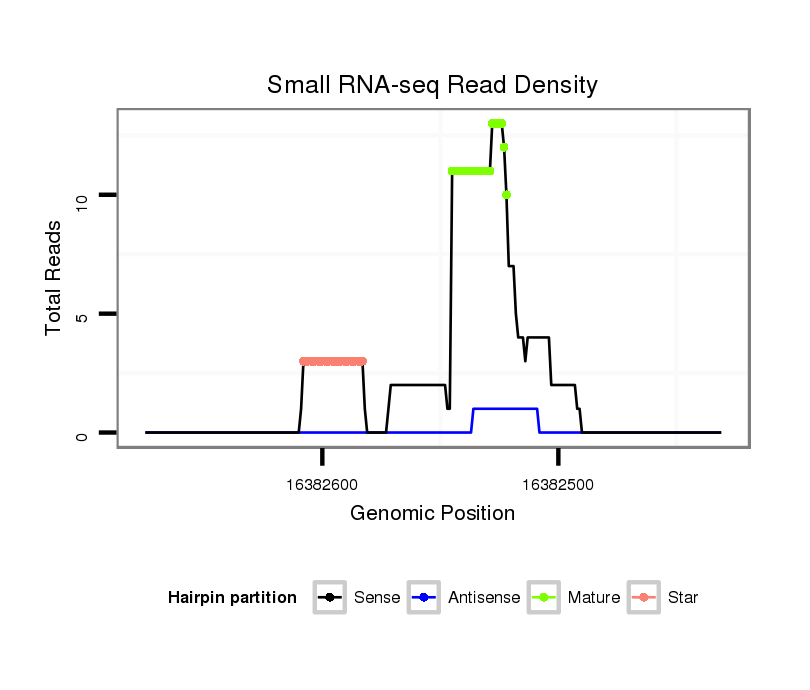
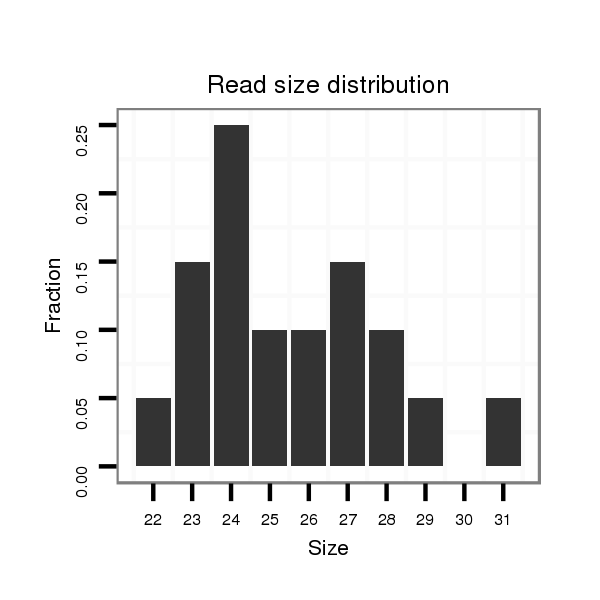
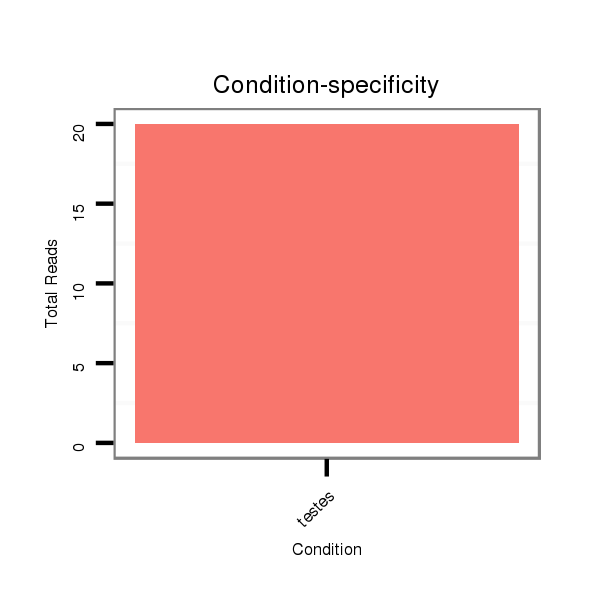
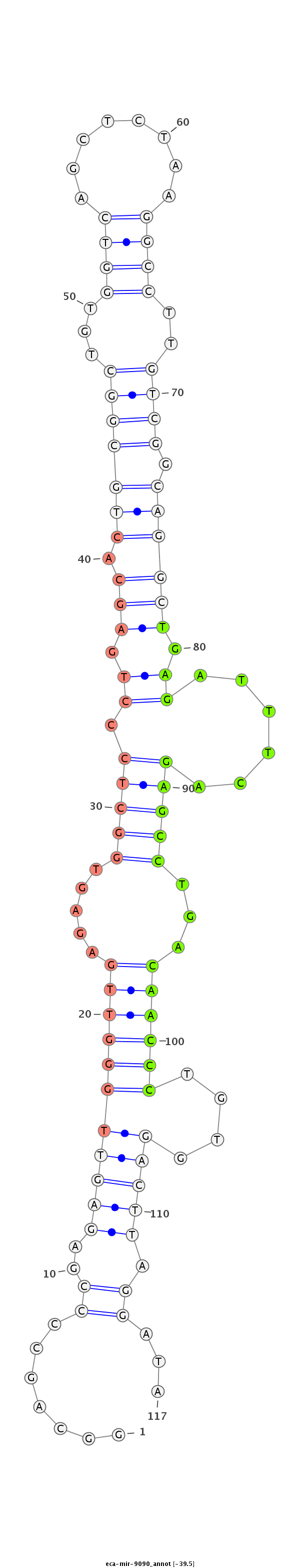
ID:eca-mir-9090 |
Coordinate:chr10:16382481-16382625 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -39.5 | -38.8 | -38.8 |
 |
 |
 |
|
CTCACCACGGGCTCTGAGGAGCCGGAATGTTCCGGTGGAGTCTGCCAAGCCAGGCAGCCCCGAGAGTTGGGTTGAGAGTGGCTCCCTGAGCACTGCGGCTGTGGTCAGCTCTAAGGCCTTGTCGGCAGGCTGAGATTTCAGAGCCTGACAACCCTGTGGACTTAGGATAGCAGAGACCCACCTCTCCCCGCATCCATCTCCCCCTCCGCCTTCCTCCACTTCCTCCAACCCAAAGCGGGGCCCCC
****************************************************.......((..(((((((((((.....(((((.((.(((.(((((((...((((........))))..))).))))))).))......)))))...))))))....))))).))...**************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACCC........................................................................................... | 24 | 0 | 1 | 4.00 | 4 | 4 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACCCTGTGGTT.................................................................................... | 31 | 2 | 1 | 2.00 | 2 | 2 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACCCTGT........................................................................................ | 27 | 0 | 1 | 2.00 | 2 | 2 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACC............................................................................................ | 23 | 0 | 1 | 2.00 | 2 | 2 |
| ...................................................................................................................................................ACAACCCTGTGGACTTAGGATAGCA......................................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 |
| ...................................................................TGGGTTGAGAGTGGCTCCCTGAGCAC........................................................................................................................................................ | 26 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................................GTCAGCTCTAAGGCCTTGTCGGCAGGC................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................TTGGGTTGAGAGTGGCTCCCTGAGCACT....................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACCCTGTG....................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAAACCTGTGGAC.................................................................................... | 31 | 1 | 1 | 1.00 | 1 | 1 |
| .......................................................................................................................................................CCCTGTGGACTTAGGATAGCAGAGACCT.................................................................. | 28 | 1 | 1 | 1.00 | 1 | 1 |
| ........................................................................................................TCAGCTCTAAGGCCTTGTCGGCAG..................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAACCCTGTGGAC.................................................................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGAGATTTCAGAGCCTGACAAC............................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGAGATTTCAGAGCGTAACAACCCTGTGGAC.................................................................................... | 31 | 2 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................................................TGTGGACTTAGGATAGCAGAGACCCACCT.............................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................................................TAGGATAGCAGAGACCCACCTCT............................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 |
|
GAGTGGTGCCCGAGACTCCTCGGCCTTACAAGGCCACCTCAGACGGTTCGGTCCGTCGGGGCTCTCAACCCAACTCTCACCGAGGGACTCGTGACGCCGACACCAGTCGAGATTCCGGAACAGCCGTCCGACTCTAAAGTCTCGGACTGTTGGGACACCTGAATCCTATCGTCTCTGGGTGGAGAGGGGCGTAGGTAGAGGGGGAGGCGGAAGGAGGTGAAGGAGGTTGGGTTTCGCCCCGGGGG
****************************************************************************.......((..(((((((((((.....(((((.((.(((.(((((((...((((........))))..))).))))))).))......)))))...))))))....))))).))...**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...........................................................................................................................................TCTCGGACTGTTGGGACACCTGAATCCT.............................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:33 AM