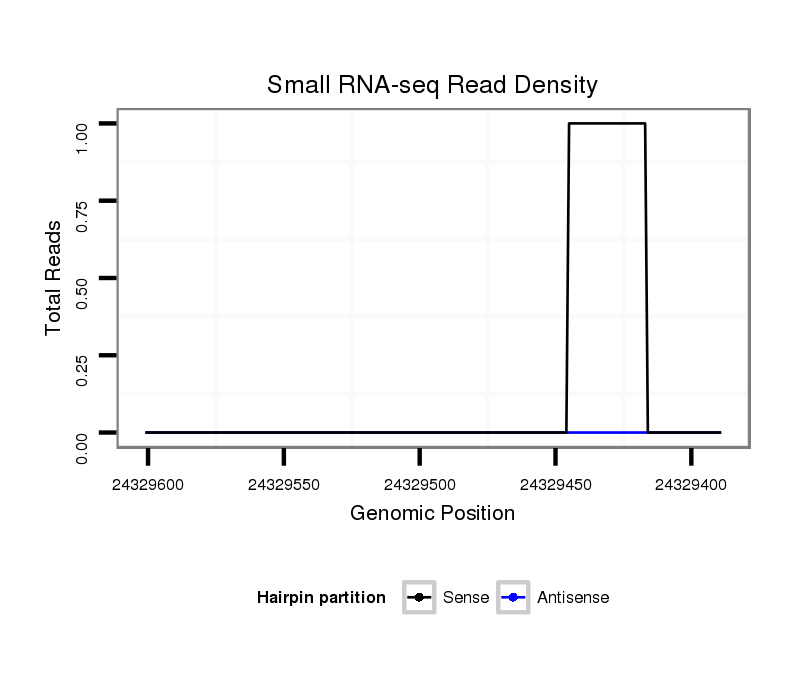
ID:eca-mir-9086 |
Coordinate:chr10:24329439-24329551 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
|
GTTGGGTCTGAGGGAGGAGGGGGCTGGGGGCCTGGACTCAGATCCTGAAGGGGCTGGATCCTGAGTCCCGGGCCCCTTGTCTGACCTCTCTCCCTGCAGGGTGTGACTCTGACGGACCTGAAGGAAGCAGAGAAGGCCGCAGGGAAGGCCCCAGAGCCAGAAAAGTCGGGCCTGCAGAGCCTGGTGAGAGGGGTCAGGTGATCGGGCAGTGGG
***********************************.......((......(((((.(((.(((..((..(((((((((((((...((((((.(((.((((.(((.......)))..)))).))).)).))))..))..))))))..)))))..))..)))....))).)))))...))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ............................................................................TTGTCTGACCTCTCTCCCTGCAGAA................................................................................................................ | 25 | 2 | 1 | 3.00 | 3 | 3 |
| ............................................................................................CACACAGGGTGTGACTCTGACGGACCT.............................................................................................. | 27 | 3 | 1 | 2.00 | 2 | 2 |
| .............................................................................................ACACAGGGTGTGACTCTGACGGACCTGAAGG......................................................................................... | 31 | 3 | 1 | 2.00 | 2 | 2 |
| ...........................................................................CTTGTCTGACCTCTCTCCCTGCAGAG................................................................................................................ | 26 | 1 | 1 | 2.00 | 2 | 2 |
| ...........................................................................CTTGTCTGACCTCTCTCCCTGCAGA................................................................................................................. | 25 | 1 | 1 | 2.00 | 2 | 2 |
| ............................................................................TTGTCTGACCTCTCTCCCTGCAGA................................................................................................................. | 24 | 1 | 1 | 2.00 | 2 | 2 |
| .........................................................................................................................................................GAGCCAGAAAAGTCGGGCCTGCAGAGCCTGT............................. | 31 | 1 | 1 | 2.00 | 2 | 2 |
| ...........................................................................................................................................................GCCAGAAAAGTCGGGCCTGCAGAGCCTGGAT........................... | 31 | 2 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................TGACGGACCTGAAGGAAGCAGAGAAA.............................................................................. | 26 | 1 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................................................AGCCAGAAAAGTCGGGCCTGCAGAGCCTGTT............................ | 31 | 1 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................TGACGGACCTGGAGGAAGCAGAGAAG.............................................................................. | 26 | 1 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................TGACGGACCTGAAGGAAGCAGAGAAGT............................................................................. | 27 | 1 | 1 | 1.00 | 1 | 1 |
| ............................................................................TTGTCTGACCTCTCTCCCTGCAGAGA............................................................................................................... | 26 | 2 | 1 | 1.00 | 1 | 1 |
| ............................................................................................CACACAGGGTGTGACTCTGACGGACC............................................................................................... | 26 | 3 | 1 | 1.00 | 1 | 1 |
| ............................................................................................................................................................CCAGAAAAGTCGGGCCTGCAGAGCCTGGT............................ | 29 | 0 | 1 | 1.00 | 1 | 1 |
| ...........................................................................CTTGTCTGACCTCTCTCCCTGCAGAA................................................................................................................ | 26 | 2 | 1 | 1.00 | 1 | 1 |
| ............................................................................ATGTCTGACCTCTCTCCCTGCAGAA................................................................................................................ | 25 | 3 | 2 | 0.50 | 1 | 1 |
|
CAACCCAGACTCCCTCCTCCCCCGACCCCCGGACCTGAGTCTAGGACTTCCCCGACCTAGGACTCAGGGCCCGGGGAACAGACTGGAGAGAGGGACGTCCCACACTGAGACTGCCTGGACTTCCTTCGTCTCTTCCGGCGTCCCTTCCGGGGTCTCGGTCTTTTCAGCCCGGACGTCTCGGACCACTCTCCCCAGTCCACTAGCCCGTCACCC
***********************************.......((......(((((.(((.(((..((..(((((((((((((...((((((.(((.((((.(((.......)))..)))).))).)).))))..))..))))))..)))))..))..)))....))).)))))...))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:34 AM