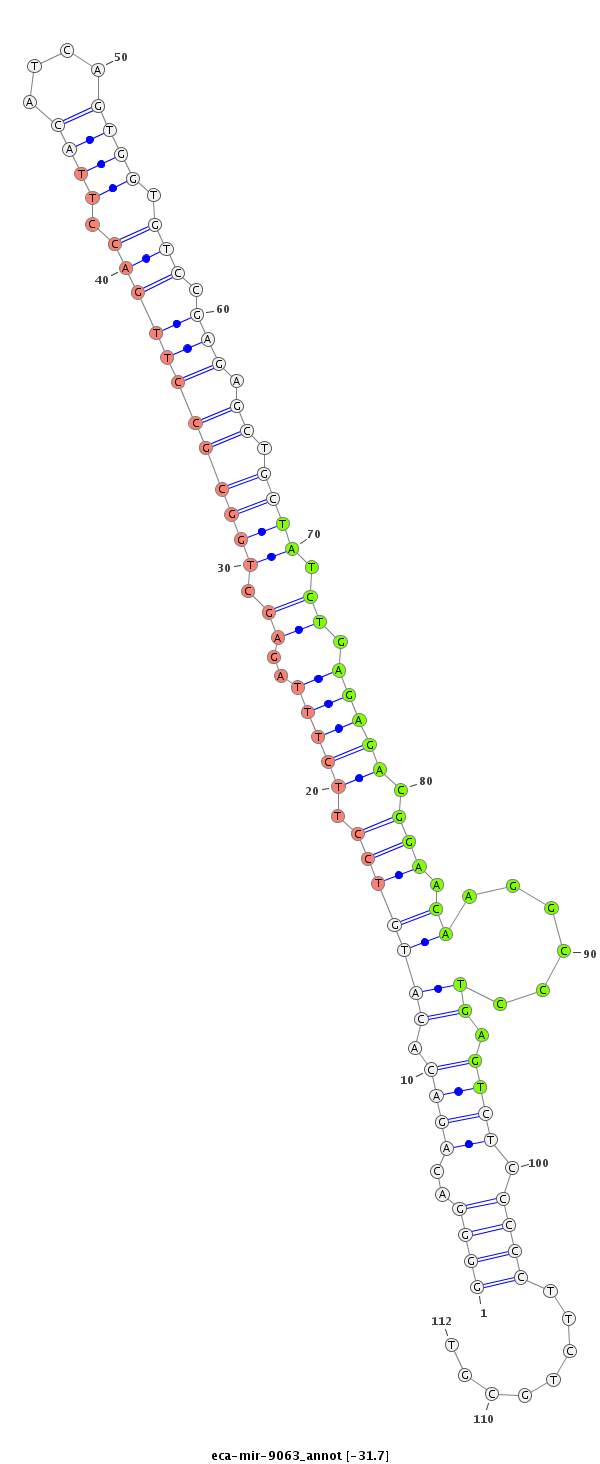
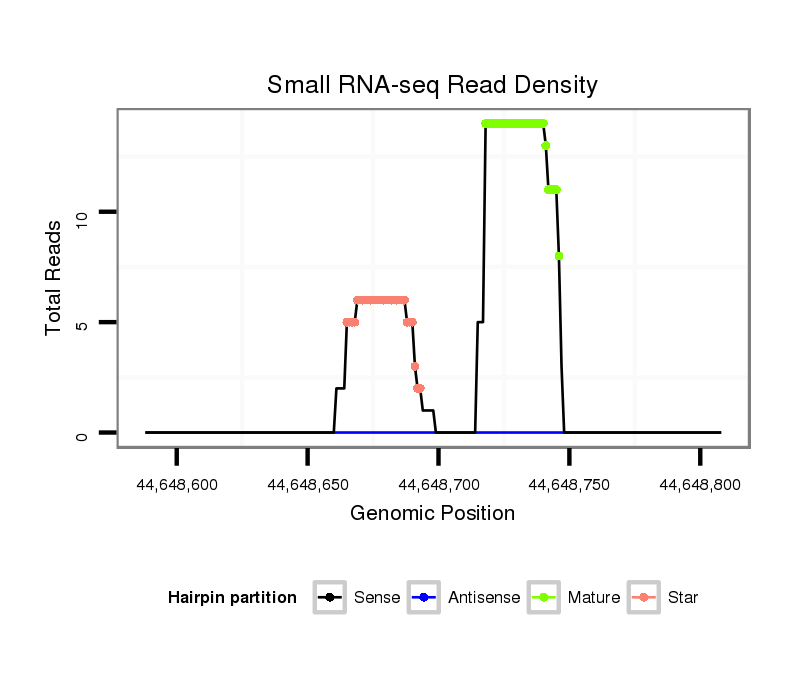
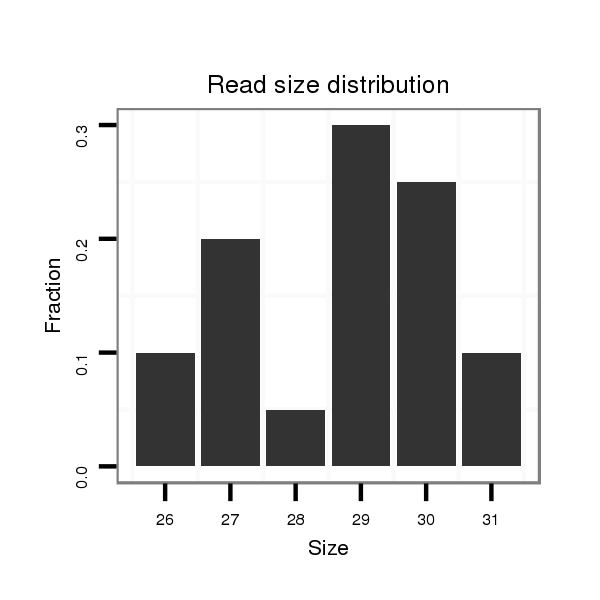
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAGT.............................................................. |
29 |
0 |
1 |
5.00 |
5 |
5 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAGTC............................................................. |
30 |
0 |
1 |
3.00 |
3 |
3 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAGA.............................................................. |
29 |
1 |
1 |
3.00 |
3 |
3 |
| ...............................................................................................................................TGCTATCTGAGAGACGGAACAAGGCCC................................................................... |
27 |
0 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................................TGCTATCTGAGAGACGGAACAAGGCCCTGAG............................................................... |
31 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................................................................................TCCGAGAGCTGCTATCTGAGAGAA............................................................................... |
24 |
1 |
1 |
2.00 |
2 |
2 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAGTTTT........................................................... |
32 |
2 |
1 |
2.00 |
2 |
2 |
| .............................................................................TCCTTCTTTAGAGCTGGCGCCTTGACCTT................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................................TTCTTTAGAGCTGGCGCCTGGACCTTACA................................................................................................................ |
29 |
1 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................................TGCTATCTGAGAGACGGAACAAGGCCT................................................................... |
27 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................TCTTTAGAGCTGGCGCCTTGACCTTACATC.............................................................................................................. |
30 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................................TTCTTTAGAGCTGGCGCCTTGACCTTACT................................................................................................................ |
29 |
1 |
1 |
1.00 |
1 |
1 |
| ......................................................................CCACATGTCCTTCTTTAGAGCTGGCGCC........................................................................................................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
| .........................................................................CATGTCCTTCTTTAGAGCTGGCGCCTT......................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................................TGCTATCTGAGAGACGGAACAAGGCC.................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAG............................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................CATGTCCTTCTTTAGAGCTGGCGCCTTGAC...................................................................................................................... |
30 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAAGCCCTGAGA.............................................................. |
29 |
2 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................................................TCTGAGAGACGGAACAAGGCCCTGAGAA............................................................. |
28 |
2 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TATCTGAGAGACGGAACAAGGCCCTGAGTA............................................................. |
30 |
1 |
1 |
1.00 |
1 |
1 |
| .............................................................................TCCTTCTTTAGAGCTGGCGCCTTGACC..................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ...................................................................................TTTAGAGCTGGCGCCTTGACCTTACAAA.............................................................................................................. |
28 |
2 |
1 |
1.00 |
1 |
1 |
| .............................................................................TCCTTCTTTAGAGCTGGCGCCTTGAC...................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TATCTGAGAGAAGGAACAAGGCCCTGAGTCT............................................................ |
31 |
1 |
1 |
1.00 |
1 |
1 |