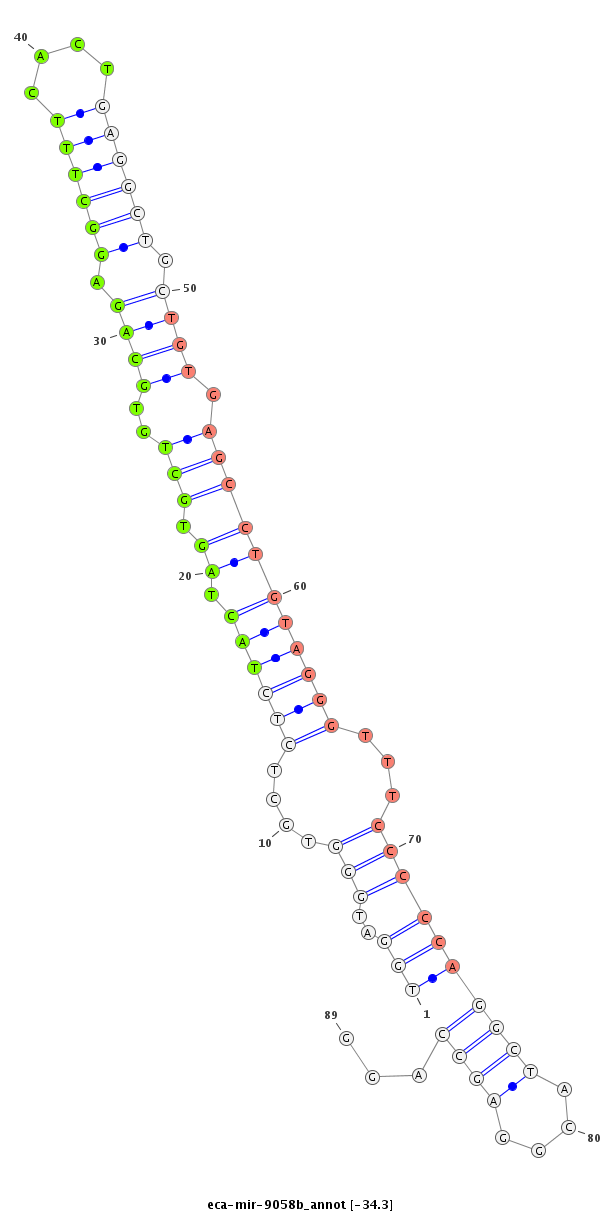
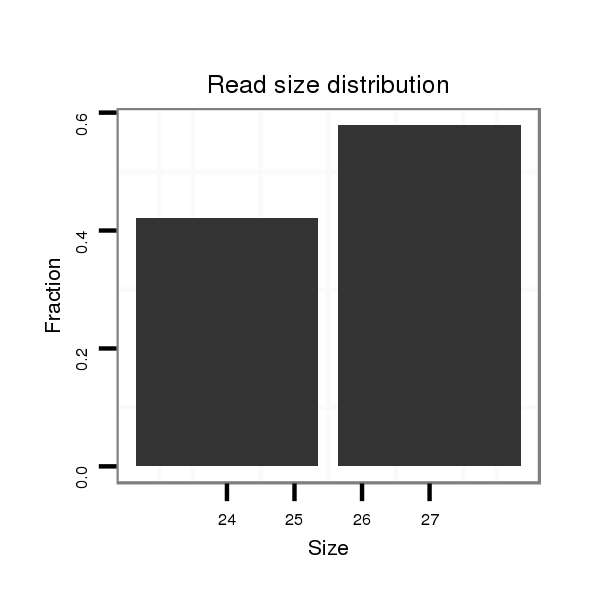
ID:eca-mir-9058b |
Coordinate:chrX:42363578-42363710 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -34.1 | -34.1 | -33.9 |
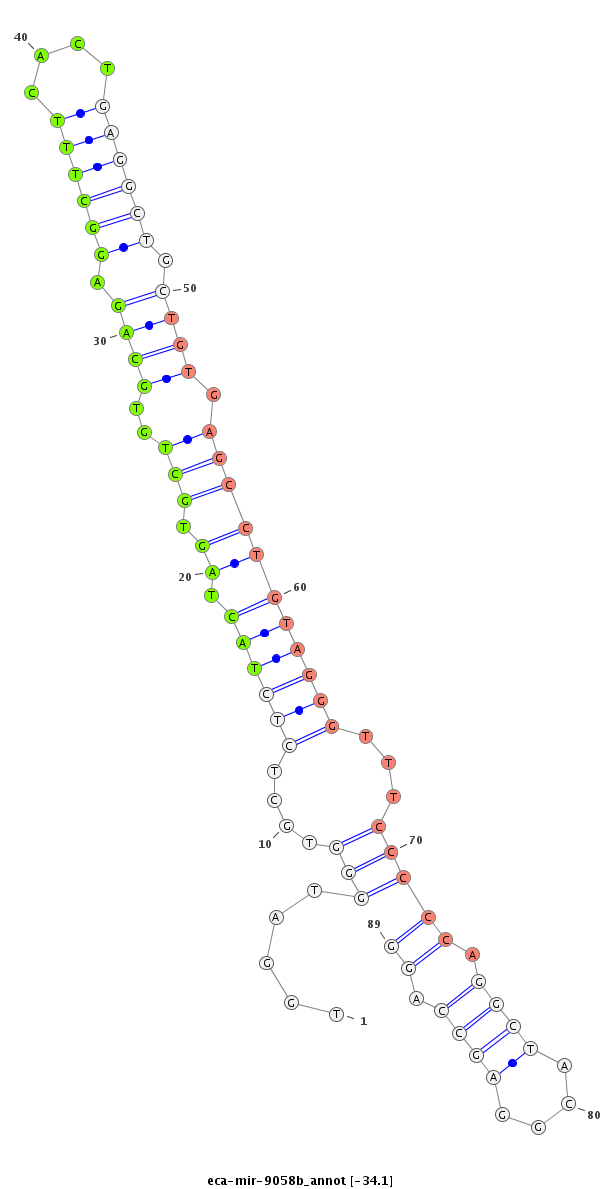 |
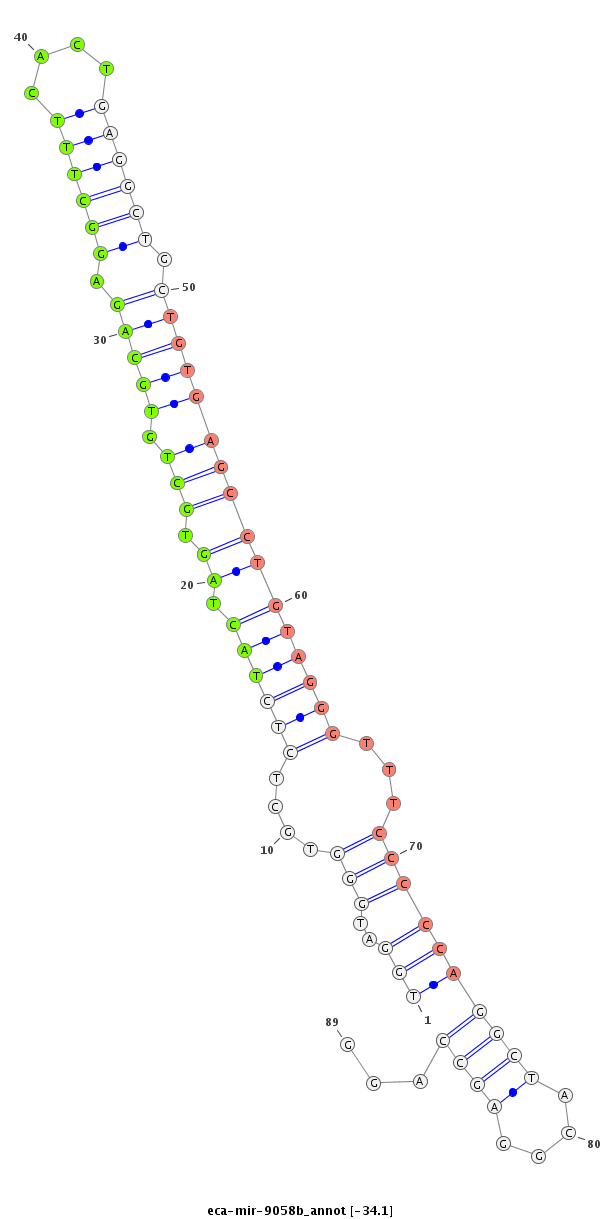 |
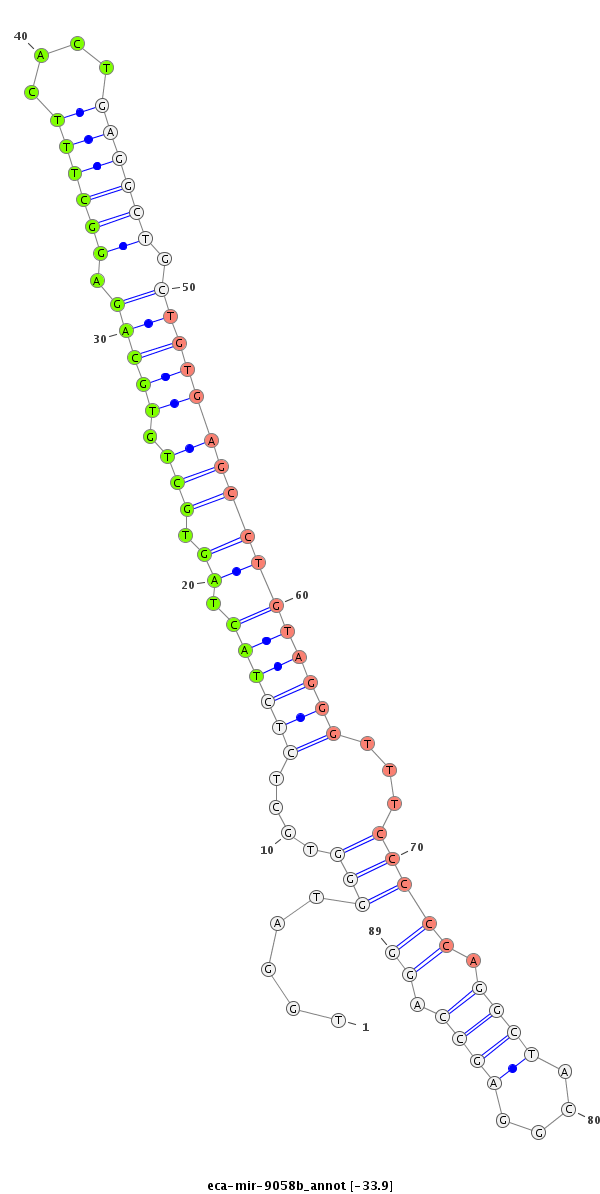 |
|
CATCTTCTACAGTCTGTGGTTAGCTCCCCTAGCTATGCTACTTTTGTCCCGGGGTCTTCCAGCCTTGTGGCTGCTGGATGGGTGCTCTCTACTAGTGCTGTGCAGAGGCTTTCACTGAGGCTGCTGTGAGCCTGTAGGGTTTCCCCCAGGCTACGGAGCCAGGTCGCTGGAACGCCACCCAGCCCCAGTCCTGTTCCCCAGGAACTCGGGGAGCCCTTTGCCCTGACTGGGGA
**************************************************************************(((..(((....((((((.((.(((..((((.((((((....)))))).)))).)))))))))))...))))))((((....))))...********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .........................................................................................TACTAGTGCTGTGCAGAGGCTTTCACT..................................................................................................................... | 27 | 0 | 20 | 0.25 | 5 | 5 |
| ............................................................................................................................TGTGAGCCTGTAGGGTTTCCCCCA..................................................................................... | 24 | 0 | 11 | 0.18 | 2 | 2 |
| ....TTCTACAGTCTGTGGTTAGCTCCCCTAGC........................................................................................................................................................................................................ | 29 | 0 | 14 | 0.14 | 2 | 2 |
|
GTAGAAGATGTCAGACACCAATCGAGGGGATCGATACGATGAAAACAGGGCCCCAGAAGGTCGGAACACCGACGACCTACCCACGAGAGATGATCACGACACGTCTCCGAAAGTGACTCCGACGACACTCGGACATCCCAAAGGGGGTCCGATGCCTCGGTCCAGCGACCTTGCGGTGGGTCGGGGTCAGGACAAGGGGTCCTTGAGCCCCTCGGGAAACGGGACTGACCCCT
**********************************************************************(((..(((....((((((.((.(((..((((.((((((....)))))).)))).)))))))))))...))))))((((....))))...************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .........................................................................GACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 26 | 0 | 20 | 0.55 | 11 | 11 |
| ........................................................................CGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 27 | 0 | 20 | 0.45 | 9 | 9 |
| .....................................................................CGACGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 30 | 0 | 20 | 0.20 | 4 | 4 |
| ....................................................................CCGACGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 31 | 0 | 20 | 0.15 | 3 | 3 |
| ......................................................................GACGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 29 | 0 | 20 | 0.10 | 2 | 2 |
| .......................................................................ACGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 |
| ..............................................................................................CACGACACGTCTCCGAAAGTGACTCCGAC.............................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 1 |
| ....AAGATGTCAGACACCAATCGGTG.............................................................................................................................................................................................................. | 23 | 2 | 20 | 0.05 | 1 | 1 |
| .....................................................................CGACGAGCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 30 | 1 | 20 | 0.05 | 1 | 1 |
| ...........................................................GTCGGAACACCGACGACCTACCCACGAGAGATGATCACGA...................................................................................................................................... | 40 | 0 | 20 | 0.05 | 1 | 1 |
| ...................................................................................................CACGTCTCCGAAAGTGACT................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 |
| ........................................................................CGACCTACCCACGAGAGATGATCACGAC..................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 |
| GTAGGAGATGTCAGACACCAAT................................................................................................................................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 1 |
| ....................................................................CCGACGACCTACCCACGAGAGATGATCAC........................................................................................................................................ | 29 | 0 | 20 | 0.05 | 1 | 1 |
| ..AGAAGATGTCAGACACCAAT................................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 |
| ....................................................................................................................................................................................TCGGGGTCAGGACAAGGGGTCCTTGAGC......................... | 28 | 0 | 20 | 0.05 | 1 | 1 |
| ..............................................................................ACCCACGAGAGAAGATCACGACAC................................................................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 1 |
| .......................................................................ACGACCTACCCACGAGAGATGATCACGC...................................................................................................................................... | 28 | 1 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 12:13 PM