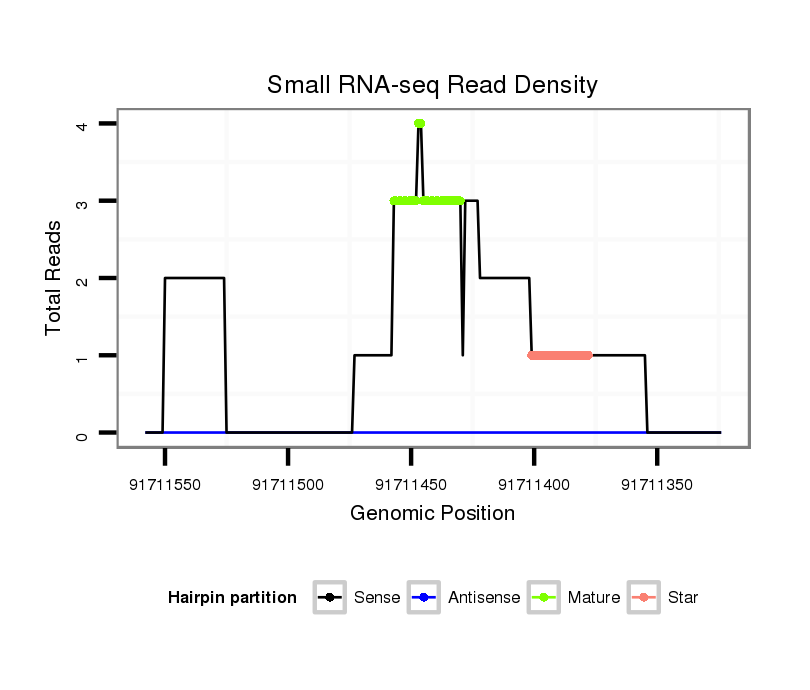
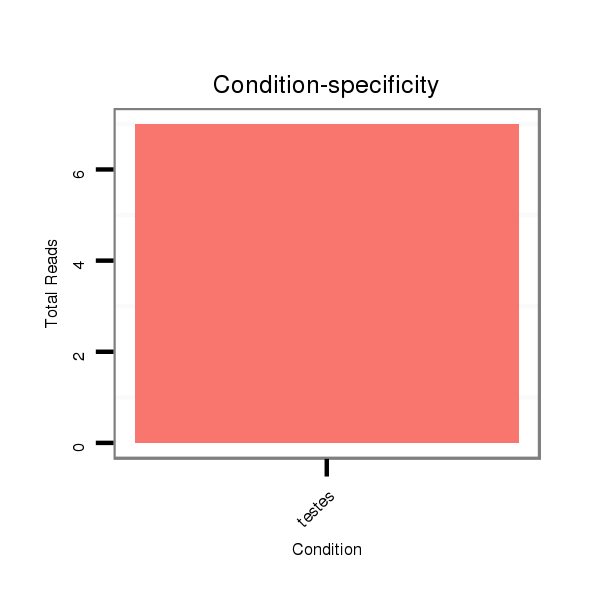
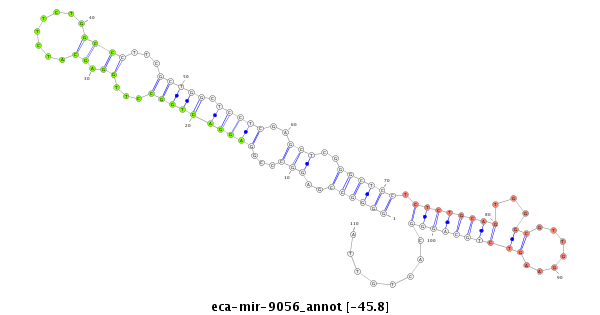
| .....................................................................................................AGGACTGGCCTTGGAGCATCTTCTGGCC.......................................................................................................... |
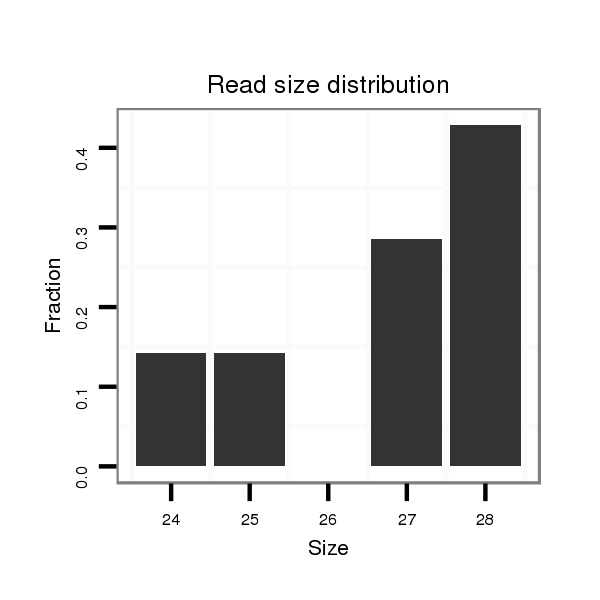
28 |
0 |
1 |
2.00 |
2 |
2 |
| ........TGTGCGAGGACTGACGTGAACGGCA.......................................................................................................................................................................................................... |
25 |
0 |
1 |
2.00 |
2 |
2 |
| ..................................................................................................................................TTCGCTGGCTCCTCGAGCTCGGGCTCC.............................................................................. |
27 |
0 |
1 |
2.00 |
2 |
2 |
| ............................................................................................................................................................................TTGGAAGTCTGCAGGGCACTGTTACA..................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................................TTCTGGCCCTTCGCTGGCTCCTCGAGA....................................................................................... |
27 |
1 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................................................................................................TGCAGGGCACTGTTACTCCAGCC............................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................................TGGGGCCGAGGCCCGGAGGACTGGCCTT.......................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TTGGAGCATCTTCTGGCCCTTCGCT................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................AGGACTGGCCTTGGAGCATATTCTGGCC.......................................................................................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
| ..............................................................................................................................................TCGAGCTCGGGCTCCTCTCTGAAGTGGGCGT.............................................................. |
31 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................................................................................................................................................TTGGAAGTCTGCAGGGCACTGTTACTT.................................... |
27 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TTCGCTGGCTCCTCGAGCTCGGGCTCCAG............................................................................ |
29 |
2 |
1 |
1.00 |
1 |
1 |
| .............................................................................................................................................................TCTCTGCAGTGGGCGTTGGAAGTC...................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |