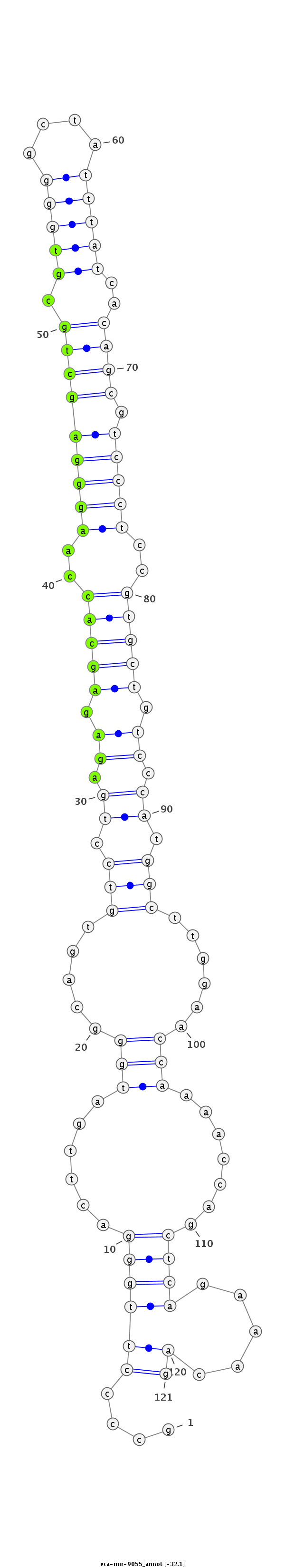
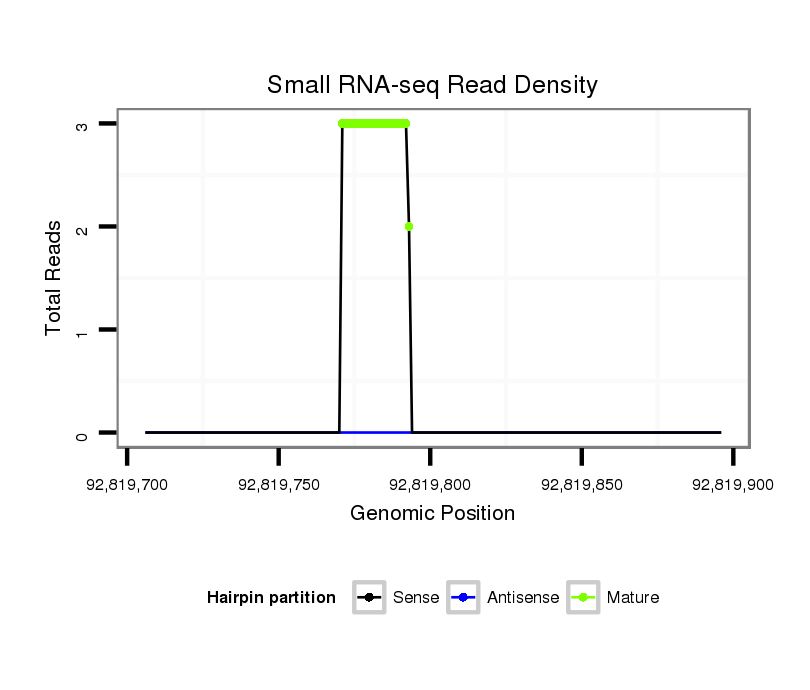
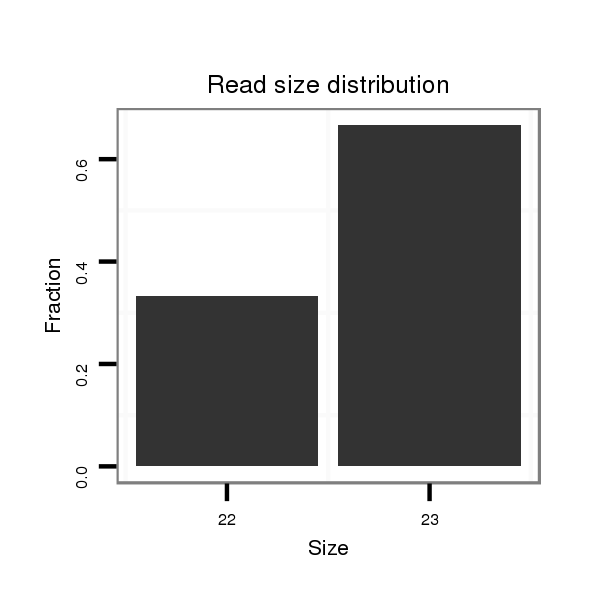
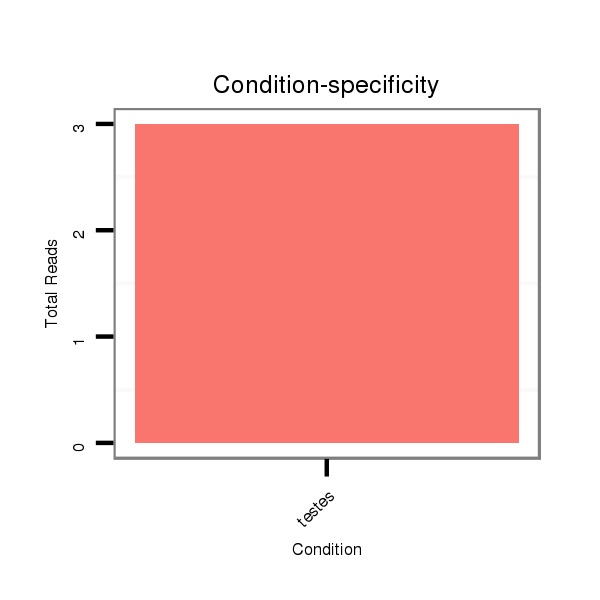
| .................................................................AGAGAGCACCAAGGGAGCTGCGAA...................................................................................................... |
24 |
2 |
1 |
3.00 |
3 |
3 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGAAT..................................................................................................... |
25 |
3 |
1 |
2.00 |
2 |
2 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGA....................................................................................................... |
23 |
1 |
1 |
2.00 |
2 |
2 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGT....................................................................................................... |
23 |
0 |
1 |
2.00 |
2 |
2 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGAAA..................................................................................................... |
25 |
3 |
1 |
2.00 |
2 |
2 |
| .................................................................AGAGAGCACCAAGGGAGCTGA......................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCG........................................................................................................ |
22 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................CGAGAGCACCAAGGGAGCTGCG........................................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCT........................................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGTATT.................................................................................................... |
26 |
3 |
1 |
1.00 |
1 |
1 |
| .................................................................CGAGAGCACCAAGGGAGCTGCGAA...................................................................................................... |
24 |
3 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGAGA..................................................................................................... |
25 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGAAT....................................................................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGAT...................................................................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGCGAAG..................................................................................................... |
25 |
2 |
1 |
1.00 |
1 |
1 |
| ........................................................................................................................TCCCGCGGCTTGGAACCAAAACCA............................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................AGAGAGCACCAAGGGAGCTGAAA....................................................................................................... |
23 |
3 |
16 |
0.13 |
2 |
2 |