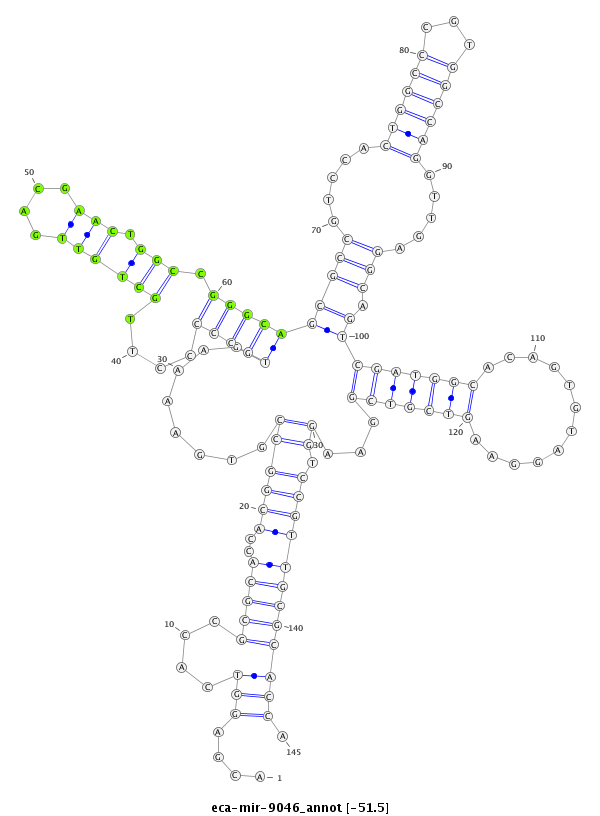
ID:eca-mir-9046 |
Coordinate:chr1:127270581-127270695 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -51.5 | -51.3 | -51.3 |
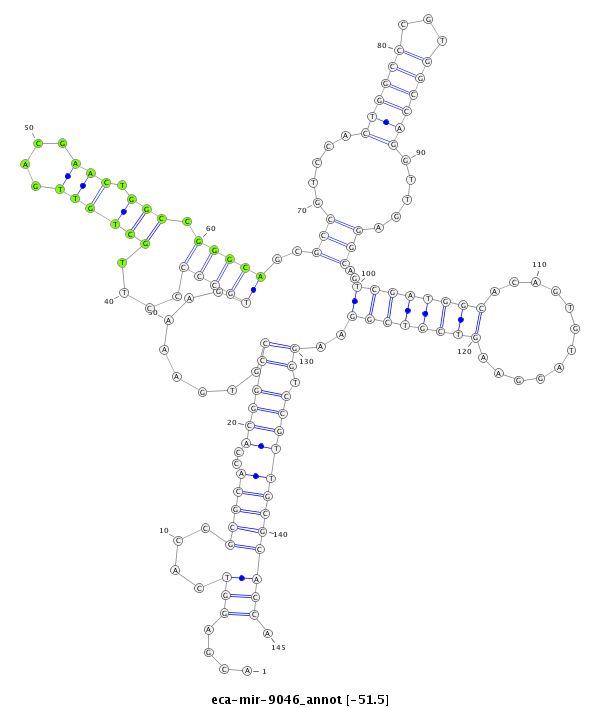 |
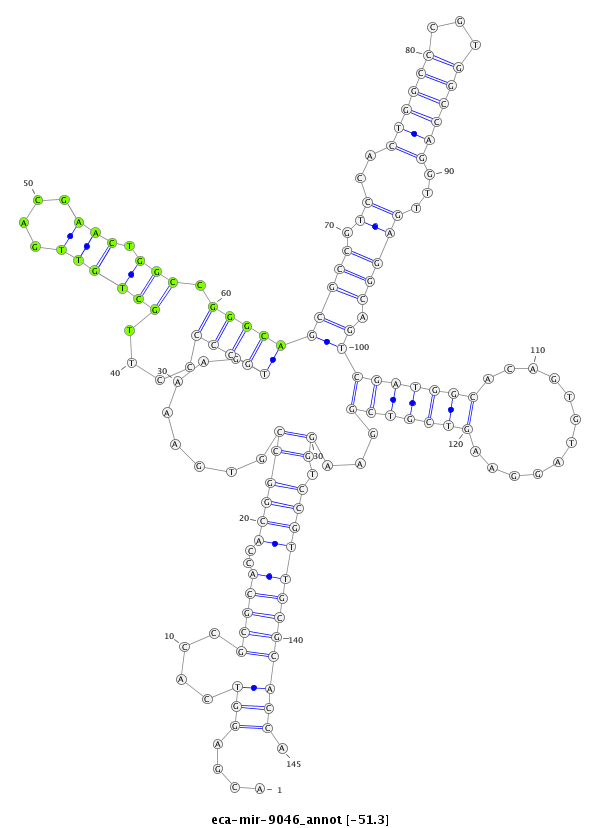 |
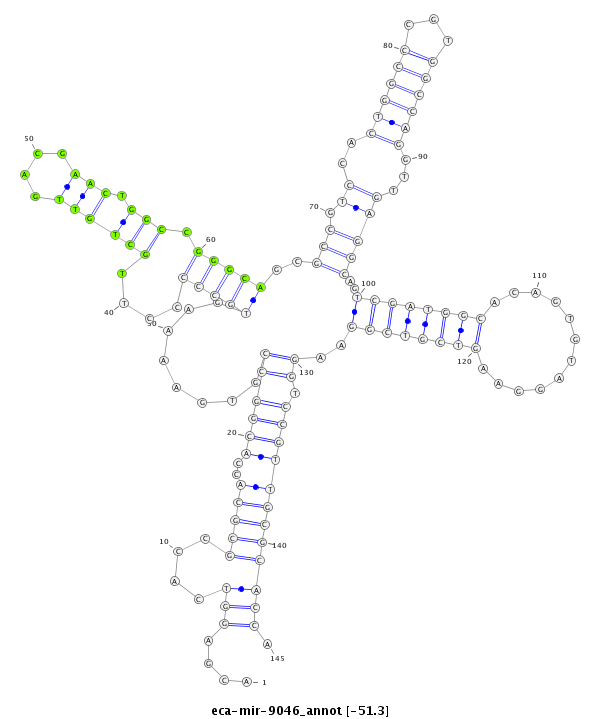 |
|
CATAGAGCAGCGTGAACATGCGGTTGACGGTATAGACGAGGTCACCGCGCACCACGGCCGTGAAAAGTGCCCCCTTGCTGTTGACGAACTGGCCGGGCAGCGCCGTCCACTGGCCCGTGGCCAGGTTGAGGCAGTCGATGGCACAGTGTAGGAAGTCGTCGGAAGGTCCGTTGCGCACCACATAGAGACTGTCGCGGTGGGCCACCATGCAGTGG
***********************************....(((....(((((..((((((........(((((....((((((....))).))).)))))(((((.....((((((...)))))).....))).))(((((((............)))))))...)).)))))))))))).*********************************** |
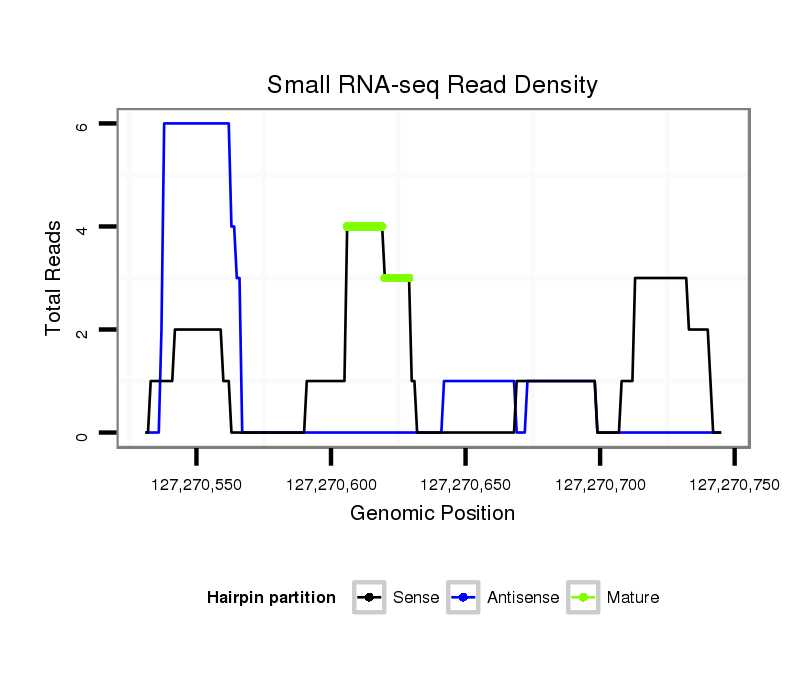
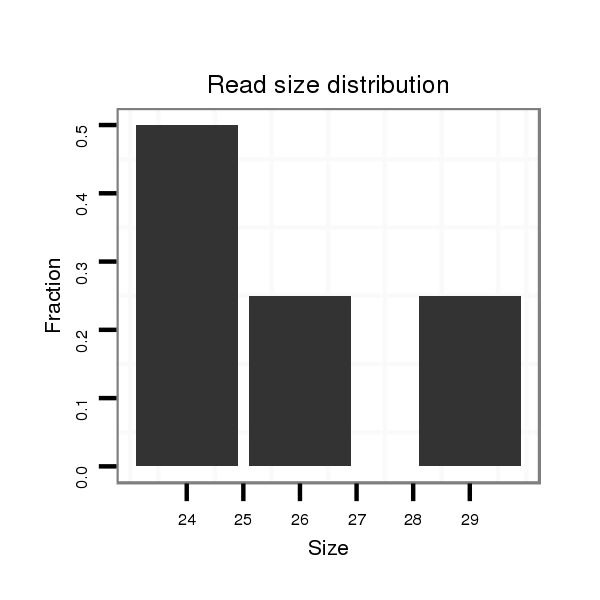
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...........................................................................TGCTGTTGACGAACTGGCCGGGCA.................................................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 |
| ...........................................................................TGCTGTTGACGAACTGGCCGGGCATA.................................................................................................................. | 26 | 2 | 1 | 2.00 | 2 | 2 |
| ...........................................................................TGCTGTTGACGAACTGGCCGGGCAGTA................................................................................................................. | 27 | 2 | 1 | 2.00 | 2 | 2 |
| .......................................................................................................................................................................CCGTTGCGCACCACATAGAGACTGTCT..................... | 27 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................................................TAGAGACTGTCGCGGTGGGCCACCATGC..... | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................................TGGCACAGTGTAGGAAGTCGTCGGAAAA................................................. | 28 | 2 | 1 | 1.00 | 1 | 1 |
| .................................................................................................................................................................................CCACATAGAGACTGTCGCGGTGGGC............. | 25 | 0 | 1 | 1.00 | 1 | 1 |
| ...........................................................................TGCTGTTGACGAACTGGCCGGGCAGC.................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ...........GTGAACATGCGGTTGACG.......................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................................................TAGAGACTGTCGCGGTGGGCCTCCATGCA.... | 29 | 1 | 1 | 1.00 | 1 | 1 |
| ..TAGAGCAGCGTGAACATGCGGTTGACGGTA....................................................................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................................................................................................................CCGTTGCGCACCACATAGAGACTGTCGTTA.................. | 30 | 3 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................................TGGCACAGTGTAGGAAGTCGTCGGAAGGTC............................................... | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................................................TAGAGACTGTCGCGGTGGGCCACCATGCA.... | 29 | 0 | 1 | 1.00 | 1 | 1 |
| ............................................................TGAAAAGTGCCCCCTTGCTGTTGACGAAC.............................................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 |
|
GTATCTCGTCGCACTTGTACGCCAACTGCCATATCTGCTCCAGTGGCGCGTGGTGCCGGCACTTTTCACGGGGGAACGACAACTGCTTGACCGGCCCGTCGCGGCAGGTGACCGGGCACCGGTCCAACTCCGTCAGCTACCGTGTCACATCCTTCAGCAGCCTTCCAGGCAACGCGTGGTGTATCTCTGACAGCGCCACCCGGTGGTACGTCACC
***********************************....(((....(((((..((((((........(((((....((((((....))).))).)))))(((((.....((((((...)))))).....))).))(((((((............)))))))...)).)))))))))))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .......GTCGCACTTGTACGCCAACTGCCATATCT................................................................................................................................................................................... | 29 | 0 | 1 | 3.00 | 3 | 3 |
| ......CGTCGCACTTGTACGCCAACTGCCAT....................................................................................................................................................................................... | 26 | 0 | 1 | 2.00 | 2 | 2 |
| ...............................................................................................................CCGGGCACCGGTCCAACTCCGTCAGCT............................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ..............................................................................................................................................TGTCACATCCTTCAGCAGCCTTCCAG............................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................................................................................CCCGGTCCAACTCCGTCAGCTACCGT........................................................................ | 26 | 1 | 1 | 1.00 | 1 | 1 |
| .......GTCGCACTTGTACGCCAACTGCCATAT..................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:32 AM