
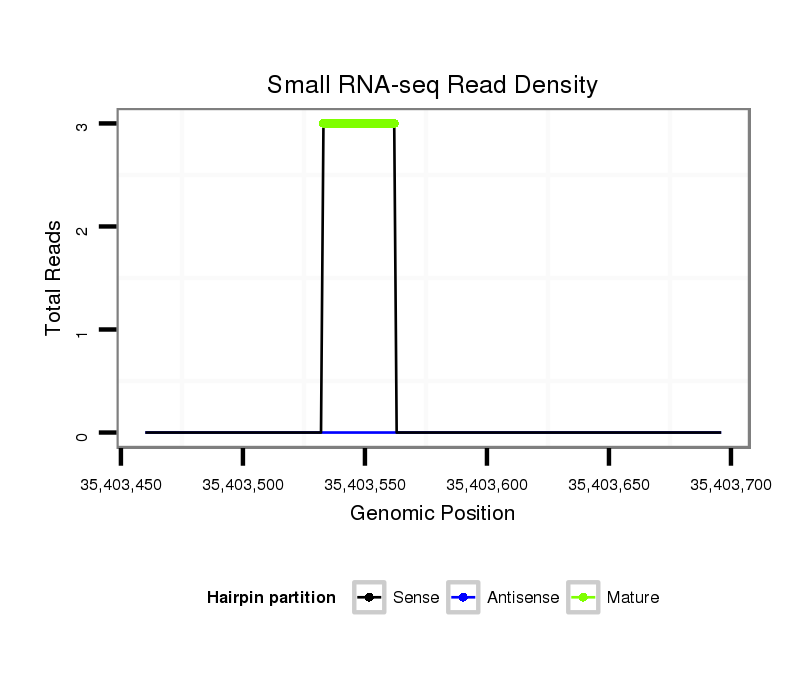
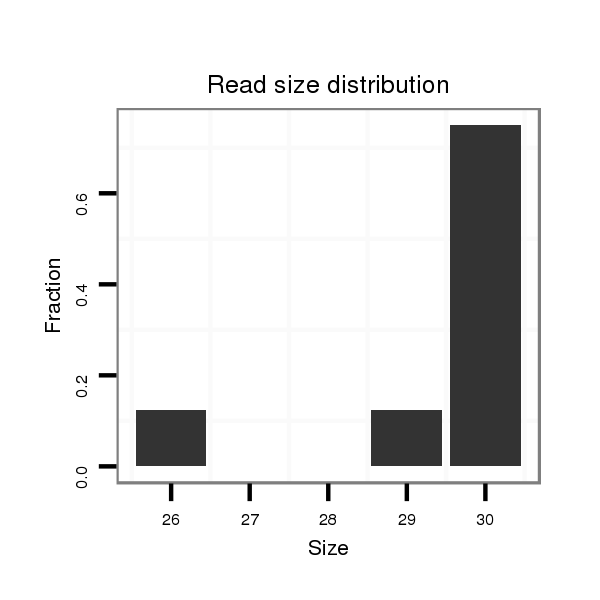
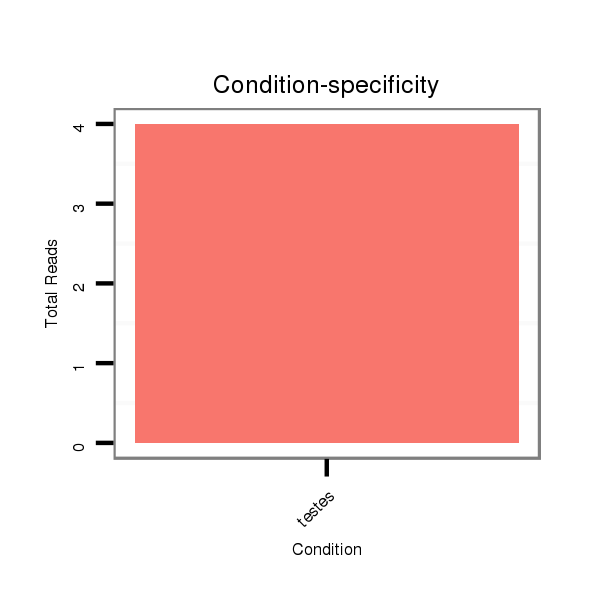
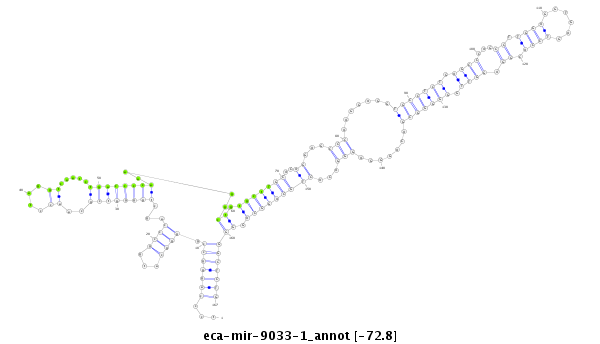
ID:eca-mir-9033-1 |
Coordinate:chr25:35403510-35403646 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -72.8 | -72.8 | -72.7 |
 |
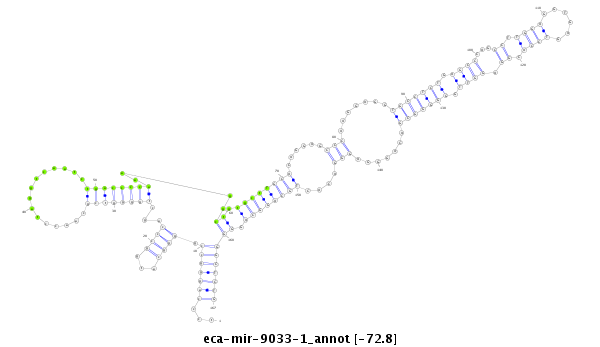 |
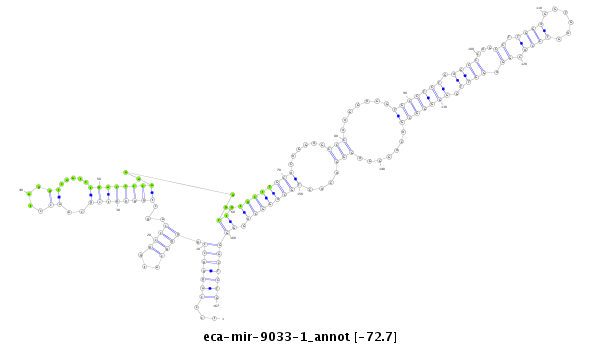 |
| mature | star |
|
tgaggcttgaagaagtcaccacatgagctggcctgtcccagggccggggcatggcccagctggggtcgcaacctaggtcaatctgaccccaaaggctgtgctTGCACAGGAGCCCAAGGAGGTGCCTCTGAGCCCAGCCTTGGAGGTGAGTCCACGGAGGTTGGGGGGCGACAGGGAGGGACACTGCAGGCAGGGGGCTCTGAGAGGGGCAAGGGCTTGGCGGGCTCCCTGAGGGAG
***********************************...(((((((.((((...))))...((((((((...............))))))))....((.(((((((..((...(((.......(((((((.((((...((.((((......)))).)).)))).))))))).......)))...))))))))).)))))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .........................................................................TAGGTCAATCTGACCCCAAAGGCTGTGCTT...................................................................................................................................... | 30 | 0 | 2 | 3.00 | 6 | 6 |
| .........................................................................TAGGTCAATCTGACCCCAAAGGCTGTGTT....................................................................................................................................... | 29 | 1 | 2 | 1.00 | 2 | 2 |
| .......................................................................................CCCAAAGGCTGTGCTTACACAGGAGCCCGAGT...................................................................................................................... | 32 | 3 | 2 | 0.50 | 1 | 1 |
| .........................................................................TAGGTCAATCTGACCCCAAAGGCTGT.......................................................................................................................................... | 26 | 0 | 2 | 0.50 | 1 | 1 |
| .................................................................................TCTGACCCCAAAGGCTGTGCTTACACAGG............................................................................................................................... | 29 | 1 | 2 | 0.50 | 1 | 1 |
| .........................................................................TAGGTCAATCTGACCCCAAAGGCTGTGCT....................................................................................................................................... | 29 | 0 | 2 | 0.50 | 1 | 1 |
|
tgTggGttgTTgTTgtGTGGTGTtgTgGtggGGtgtGGGTgggGGggggGTtggGGGTgGtggggtGgGTTGGtTggtGTTtGtgTGGGGTTTggGtgtgGtACGTGTCCTCGGGTTCCTCCACGGAGACTCGGGTCGGAACCTCCACTCAGGTGCCTCCAACCCCCCGCTGTCCCTCCCTGTGACGTCCGTCCCCCGAGACTCTCCCCGTTCCCGAACCGCCCGAGGGACTCCCTC
***********************************...(((((((.((((...))))...((((((((...............))))))))....((.(((((((..((...(((.......(((((((.((((...((.((((......)))).)).)))).))))))).......)))...))))))))).)))))))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...........................................................................................TTCCGACACGAATGTGTCCTCGGGCTCCT..................................................................................................................... | 29 | 2 | 2 | 1.00 | 2 | 2 |
| ..............................................................................................CGACACGAATGTGTCCTCGGGCTCCT..................................................................................................................... | 26 | 2 | 2 | 1.00 | 2 | 2 |
| .............................................................................................CCGACACGAATGTGTCCTCGGGCTCCT..................................................................................................................... | 27 | 2 | 2 | 1.00 | 2 | 2 |
| ............................................................................................TCCGACACGAATGTGTCCTCGGGCTCCT..................................................................................................................... | 28 | 2 | 2 | 0.50 | 1 | 1 |
| .............................................CCCCGTACCGGGTCGACCCCAGCGTTGGAT.................................................................................................................................................................. | 30 | 0 | 2 | 0.50 | 1 | 1 |
Generated: 09/01/2015 at 11:57 AM