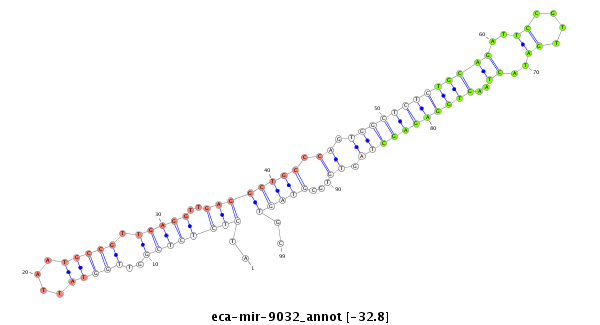
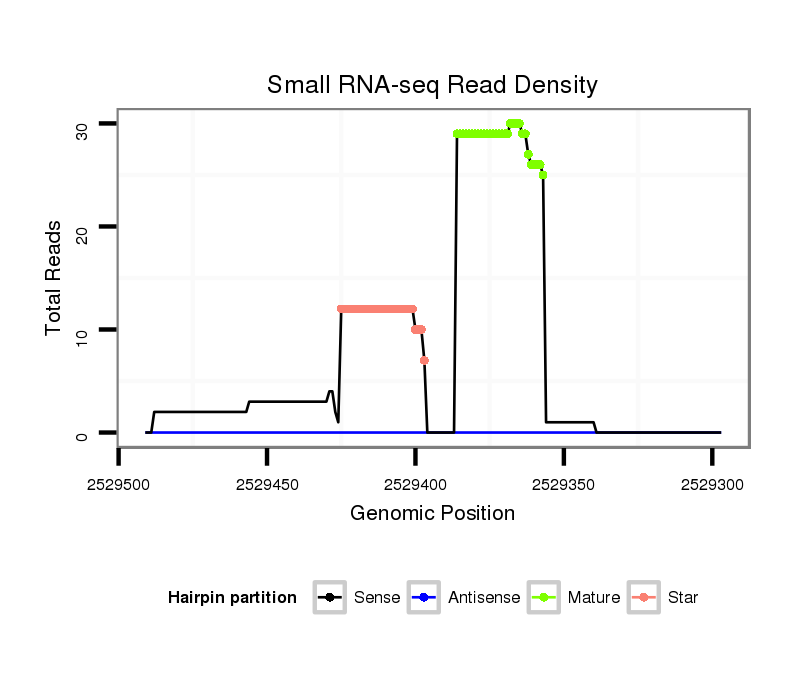
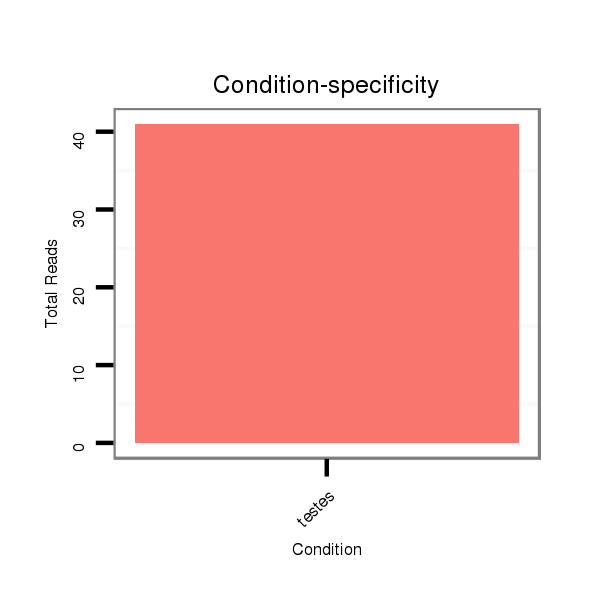
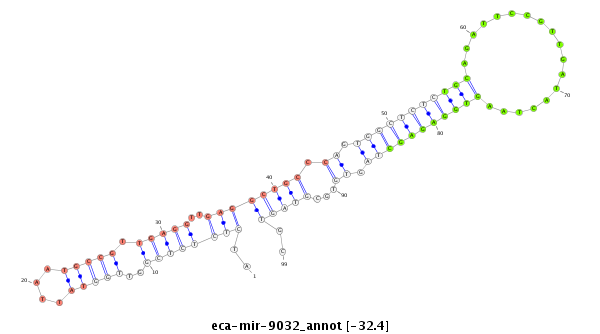
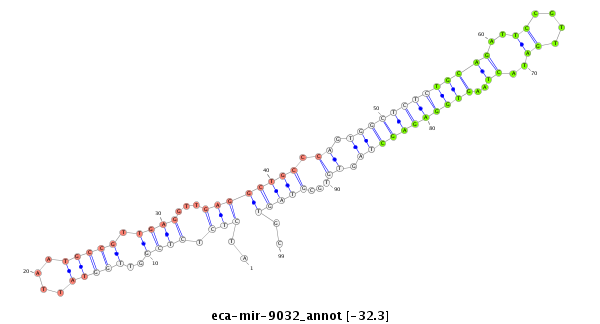
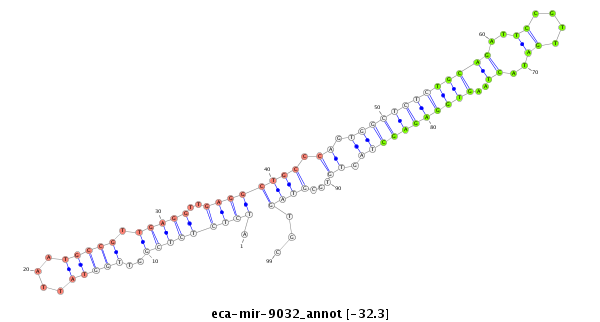
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTGGAGAGC............................................................ |
30 |
0 |
1 |
24.00 |
24 |
24 |
| ..................................................................TATTAATGCCGTTGAGGTTGAGGCTGCCC.................................................................................................... |
29 |
0 |
1 |
7.00 |
7 |
7 |
| ..................................................................TATTAATGCCGTTGAGGTTGAGGCTGCCCT................................................................................................... |
30 |
1 |
1 |
3.00 |
3 |
3 |
| ..................................................................TATTAATGCCGTTGAGGTTGAGGCTGCC..................................................................................................... |
28 |
0 |
1 |
3.00 |
3 |
3 |
| .........................................................................................CTGCCCAGTGGCTCTCTGCAGATTCCGTTAT........................................................................... |
31 |
2 |
1 |
3.00 |
3 |
3 |
| .....................................................................TAATGCCGTTGAGGTTGAGGCTGCCT.................................................................................................... |
26 |
1 |
1 |
2.00 |
2 |
2 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTG.................................................................. |
24 |
0 |
1 |
2.00 |
2 |
2 |
| ...................................TGGGACCAAAACTGAAATCTCTCTCGGTT................................................................................................................................... |
29 |
0 |
1 |
2.00 |
2 |
2 |
| ...TCGACCTCTTGAGATAAGGCCGACGCCAGGGC................................................................................................................................................................ |
32 |
0 |
1 |
2.00 |
2 |
2 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTGGAGAGT............................................................ |
30 |
1 |
1 |
2.00 |
2 |
2 |
| ..............................................................TTGGTATTAATGCCGTTGAGGTTGAGGCT........................................................................................................ |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................TTAATGCCGTTGAGGTTGAGGCTGCTT.................................................................................................... |
27 |
2 |
1 |
1.00 |
1 |
1 |
| ..................................................................TATTAATGTCGTTGAGGTTGAGGCTGCC..................................................................................................... |
28 |
1 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTGG................................................................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAG.................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTGGAGAG............................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................TATTAATGCCGTTGAGGTTGAGGCT........................................................................................................ |
25 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................................................................TGCGTTTCTGAGTGACGTGCACGCTTTCAGT..... |
31 |
1 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGGGAAGAGC............................................................ |
30 |
2 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAATTGGAGAGC............................................................ |
30 |
1 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................TGCAGATTCCGTTGATACTAAGTGGT................................................................ |
26 |
1 |
1 |
1.00 |
1 |
1 |
| ...................................TGGGACCAAAACTGAAATCTCTCTCGGTTG.................................................................................................................................. |
30 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................TAAGTGGAGAGCTAGTGTGCGTAGTGCTC........................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |