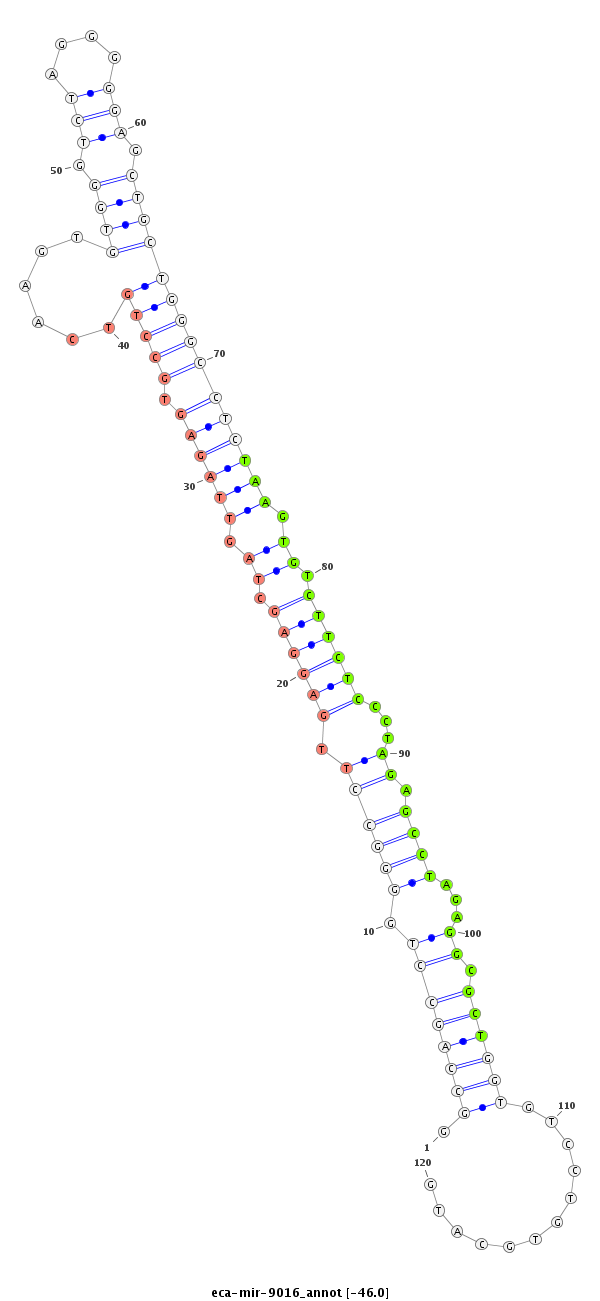
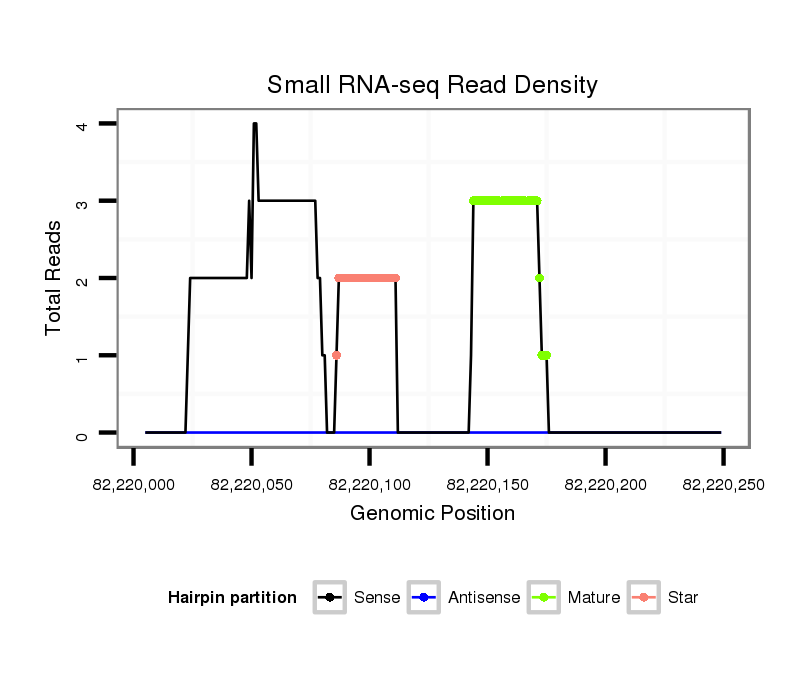
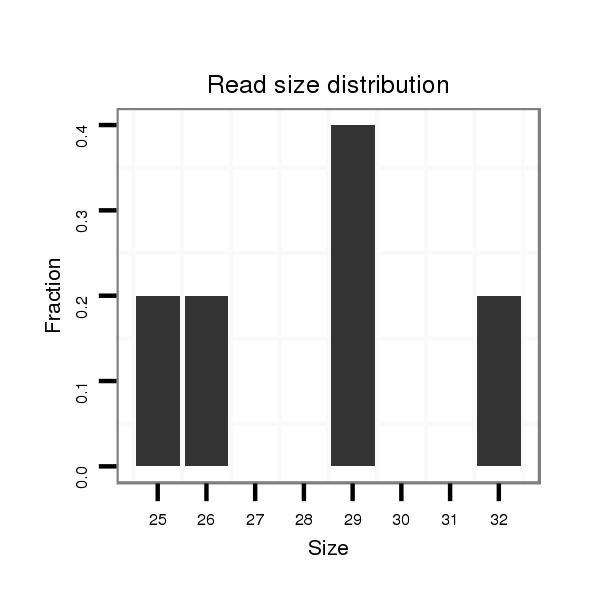
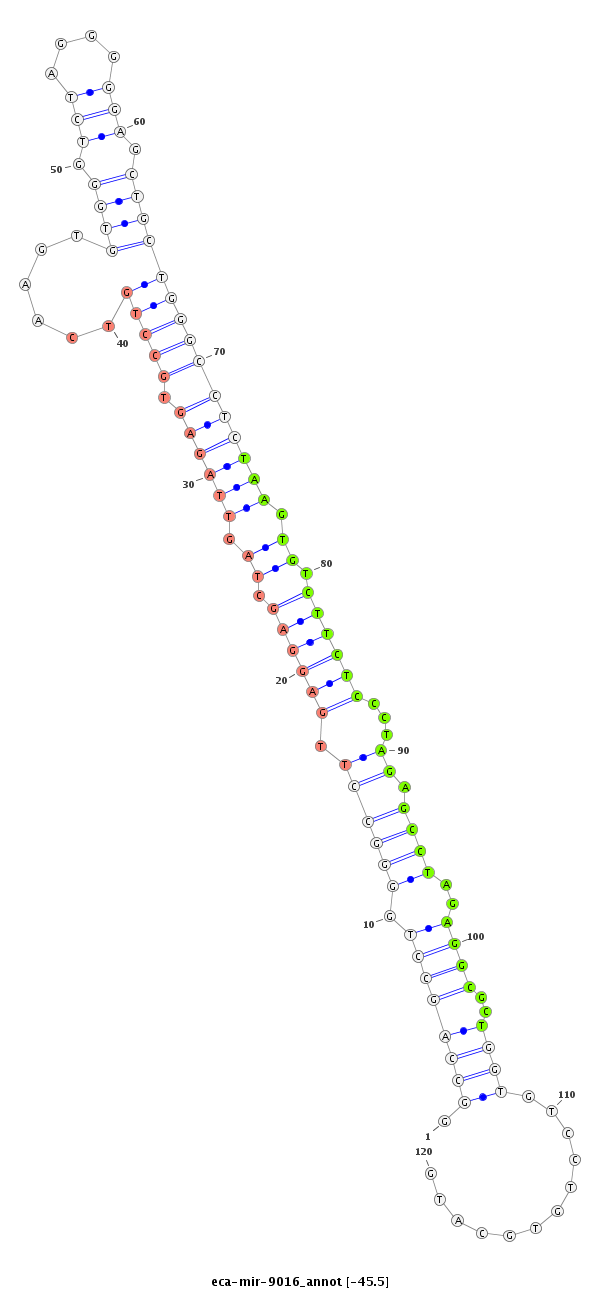
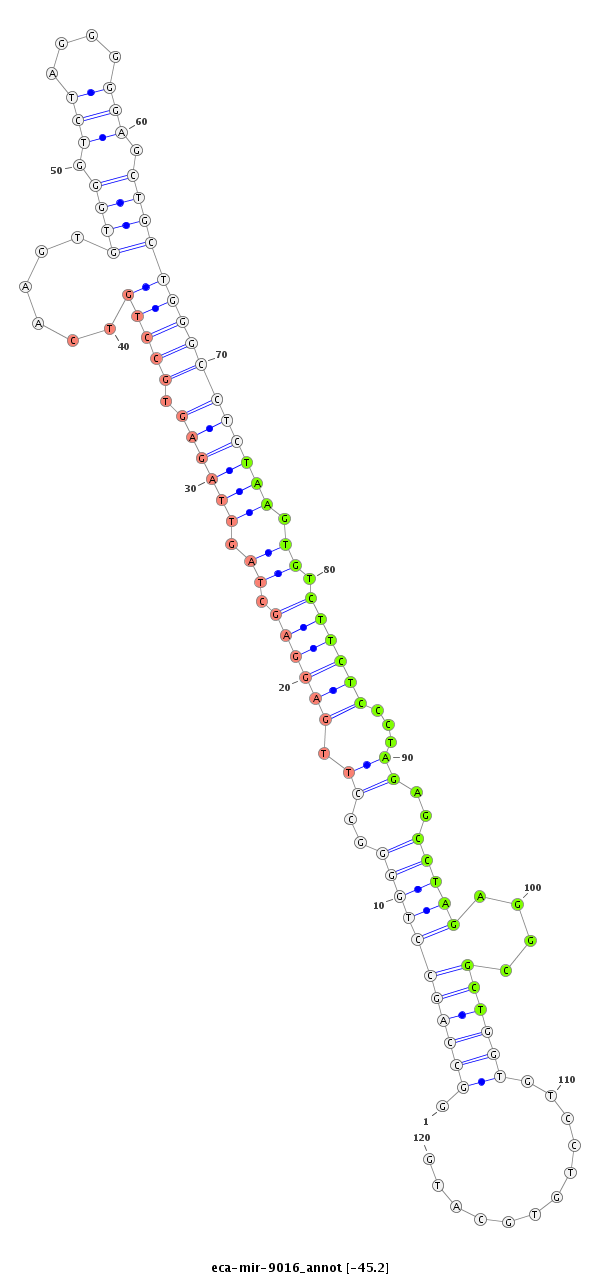
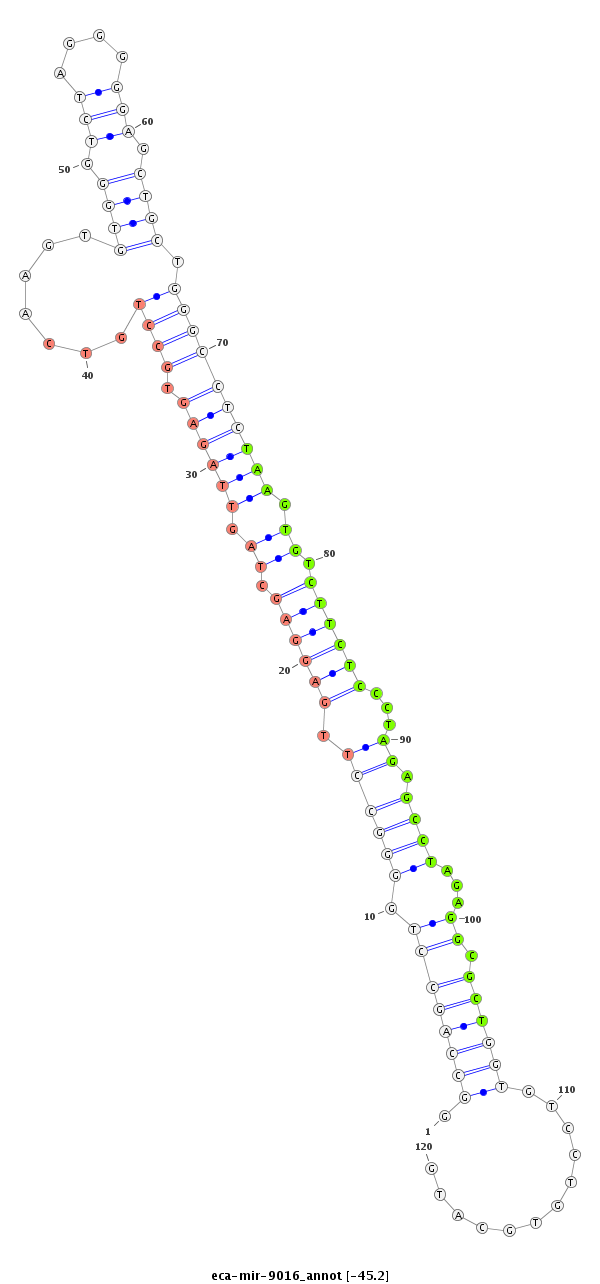
| ..............................................................................GCCTTGAGGAGCTAGTTAGAGTGTT.............................................................................................................................................. |
25 |
2 |
1 |
2.00 |
2 |
2 |
| .....................TCCCCGTGGGCCTGGAAGGGCACTGTACT................................................................................................................................................................................................... |
29 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................TGTACAAACAAGGCAGAGAACAGGCCAGCCT.......................................................................................................................................................................... |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................TAGTTAGAGTGCCTGTCAAGTGTGGGTCTT............................................................................................................................. |
30 |
1 |
1 |
1.00 |
1 |
1 |
| ..............................................TACAAACAAGGCAGAGAACAGGCCAGC............................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................................TAAGTGTCTTCTCCCTAGAGCGTAGAGGCGCT.......................................................................... |
32 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................AAGGGCACTGTACAAACAAGGCAGAGT...................................................................................................................................................................................... |
27 |
1 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................................TAAGTGTCTTCTCCCTAGAGCCTAGAGGCGCT.......................................................................... |
32 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................................TACAAACAAGGCAGAGAACAGGCCAGCCTGT........................................................................................................................................................................ |
31 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................TTGAGGAGCTAGTTAGAGTGCCTGTC.......................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................TGAGGAGCTAGTTAGAGTGCCTGTC.......................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
| ..................TTGTCCCCGTGGGCCTGGAAGGGCACT........................................................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................................TAAGTGTCTTCTCCCTAGAGCCTAGAGGC............................................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................................................................CTAAGTGTCTTCTCCCTAGAGCCTAGAGG.............................................................................. |
29 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................................TACAAACAAGGCAGAGAACAGGCCAGCCTGG........................................................................................................................................................................ |
31 |
0 |
1 |
1.00 |
1 |
1 |
| ...................TGTCCCCGTGGGCCTGGAAGGGCACTGTA..................................................................................................................................................................................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |