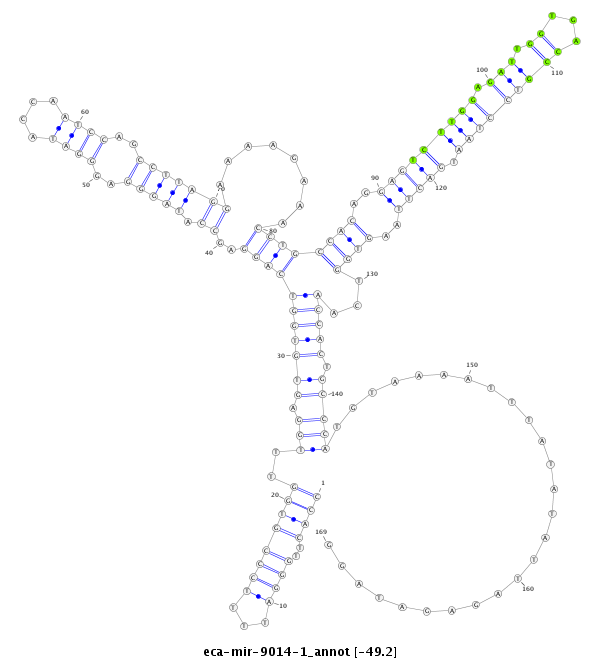



ID:eca-mir-9014-1 |
Coordinate:chr8:5879610-5879748 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
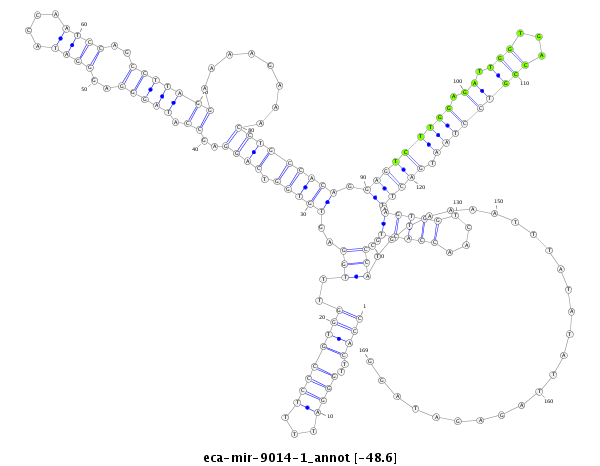
| -49.2 | -48.6 | -48.6 |
 |
 |
 |
| mature | star |
|
AGGTTAGAGTTTGGTACTTTCCTTGAACCAGAGGCCCACTTGGGATTTTCCCGTGGTTTGGAGTGTGGTCAGGAGCCATAGGGAGGGATACCAATCCAGCCTTAGGAAAAGAAACCTGCCACAGGAGTCTTGGAGATTGGTGACCGTCCTAATGACTTAAGTGGTCAACCACTGCCCATGTAAAATTTATATATTAGAGATAGGATTAGGAAGGAATGGATTAATTCATTCTTCATCAC
***********************************((((..((((...))))))))..(((.(((((((((((..((.(((((..((((....))))..)))))))........))))((((..(((((((((.(((.((...))))))))).)))))..))))...))))).)))))..........................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...............................................................................................................................TCTTGGAGATTGGTGACCG............................................................................................. | 19 | 0 | 7 | 0.14 | 1 | 1 |
| ........................................................................................................................................TTGGTGACCGTCCTAATGACTTAAGTGGT.......................................................................... | 29 | 0 | 7 | 0.14 | 1 | 1 |
| ..................................................................................................................................TGGAGATTGGTGACCGTCC.......................................................................................... | 19 | 0 | 8 | 0.13 | 1 | 1 |
| .................................................................................................................................TTGGAGATTGGTGACCGTCC.......................................................................................... | 20 | 0 | 8 | 0.13 | 1 | 1 |
| ..................................................................................................................................................................................TGTAAAATTTATATATTAGAGATAGGATT................................ | 29 | 0 | 20 | 0.10 | 2 | 2 |
| .........................................................................................................................................TGGTGACCGTCCTAATGACTTAAGTGT........................................................................... | 27 | 1 | 11 | 0.09 | 1 | 1 |
| .................................................................................................AGCCTTAGGAAAAGAAACCTGCCACAGGAGA............................................................................................................... | 31 | 1 | 13 | 0.08 | 1 | 1 |
| ............................................................................................................................................................................TGCCCATGTAAAATTTATATATTAGAGATAGT................................... | 32 | 1 | 18 | 0.06 | 1 | 1 |
|
TCCAATCTCAAACCATGAAAGGAACTTGGTCTCCGGGTGAACCCTAAAAGGGCACCAAACCTCACACCAGTCCTCGGTATCCCTCCCTATGGTTAGGTCGGAATCCTTTTCTTTGGACGGTGTCCTCAGAACCTCTAACCACTGGCAGGATTACTGAATTCACCAGTTGGTGACGGGTACATTTTAAATATATAATCTCTATCCTAATCCTTCCTTACCTAATTAAGTAAGAAGTAGTG
***********************************((((..((((...))))))))..(((.(((((((((((..((.(((((..((((....))))..)))))))........))))((((..(((((((((.(((.((...))))))))).)))))..))))...))))).)))))..........................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| TCCAATCTCAAACCATGAAAGGAACT..................................................................................................................................................................................................................... | 26 | 0 | 20 | 0.55 | 11 | 11 |
| .......................................................................CCTCGGTATCCCTCCCTATGGTTAGGT............................................................................................................................................. | 27 | 0 | 12 | 0.17 | 2 | 2 |
| ....................................................................AGTCCTCGGTTTCCCTCCCTATGGTTAGGT............................................................................................................................................. | 30 | 1 | 9 | 0.11 | 1 | 1 |
| ....................................................................AGTCCTCGGTATCCCTCCCTATGGTTAGGT............................................................................................................................................. | 30 | 0 | 9 | 0.11 | 1 | 1 |
| ....................................................CACCAAACCTCACACGCGTCCTCGGTAT............................................................................................................................................................... | 28 | 2 | 11 | 0.09 | 1 | 1 |
| .......................................................................ACTCGGTATCCCTCCCTATGGTTAGGT............................................................................................................................................. | 27 | 1 | 12 | 0.08 | 1 | 1 |
| TCCAATCTCAGAGCATGAAAGGAACTT.................................................................................................................................................................................................................... | 27 | 2 | 20 | 0.05 | 1 | 1 |
| .....................................................................................................................................TCTAACCTCTGGCAGGATTACTGAATT............................................................................... | 27 | 1 | 20 | 0.05 | 1 | 1 |
| .CCAATCTCAAACCTTGAAAGGAACTT.................................................................................................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 |
| TCCAATCTCAAACCATGAAAGGAACTTGC.................................................................................................................................................................................................................. | 29 | 1 | 20 | 0.05 | 1 | 1 |
| TCCAATCTCAAAGCATGAAAGGAACTT.................................................................................................................................................................................................................... | 27 | 1 | 20 | 0.05 | 1 | 1 |
| .CCAATCTCAGACCATGAAAGGAACTT.................................................................................................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 |
| .CCAATCTCAAACCCTGAAAGGAACTT.................................................................................................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 12:09 PM