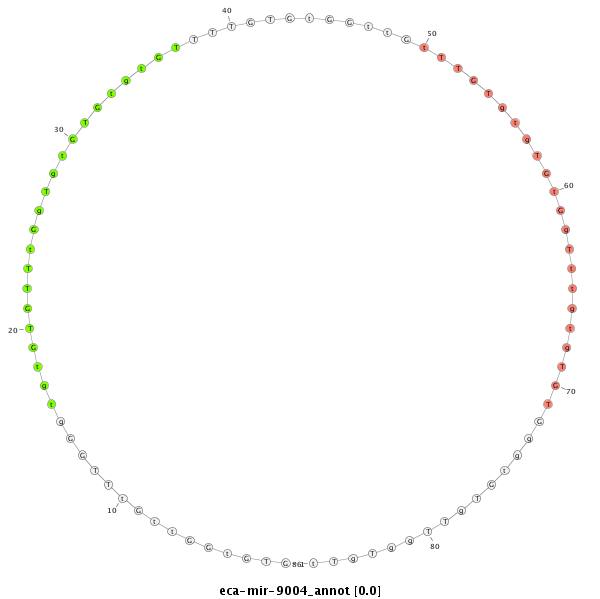
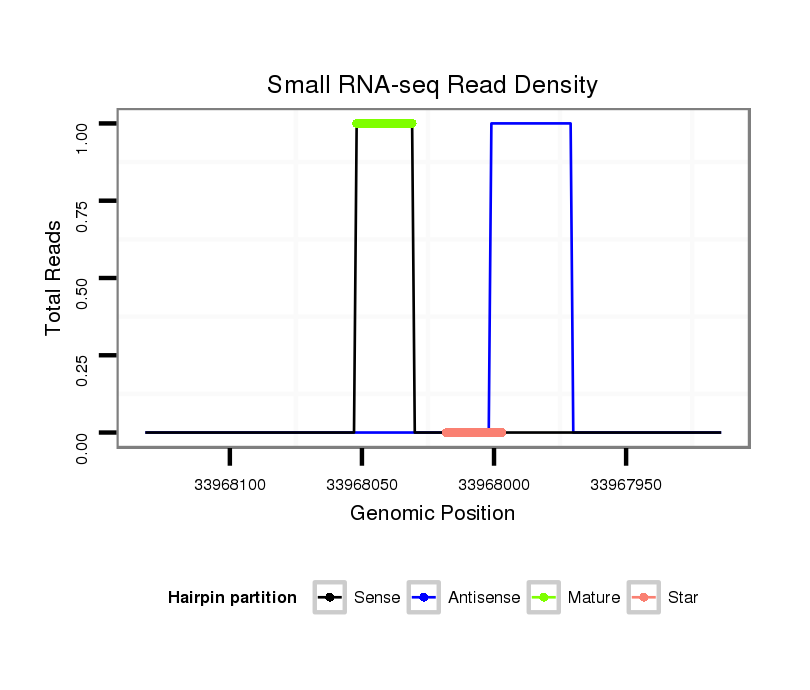
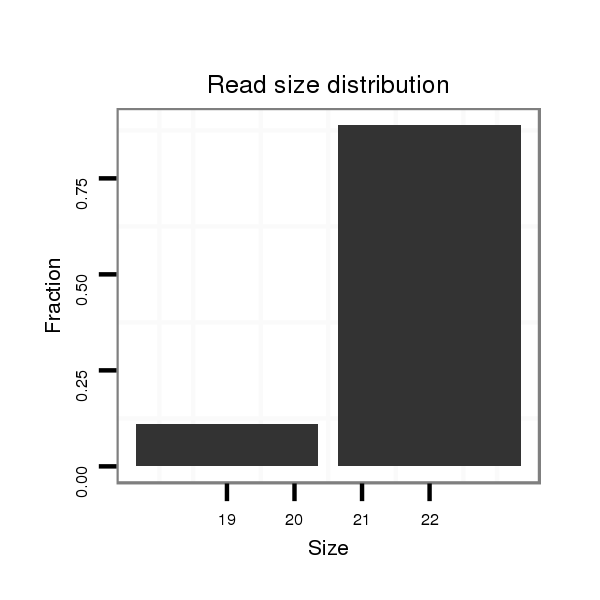
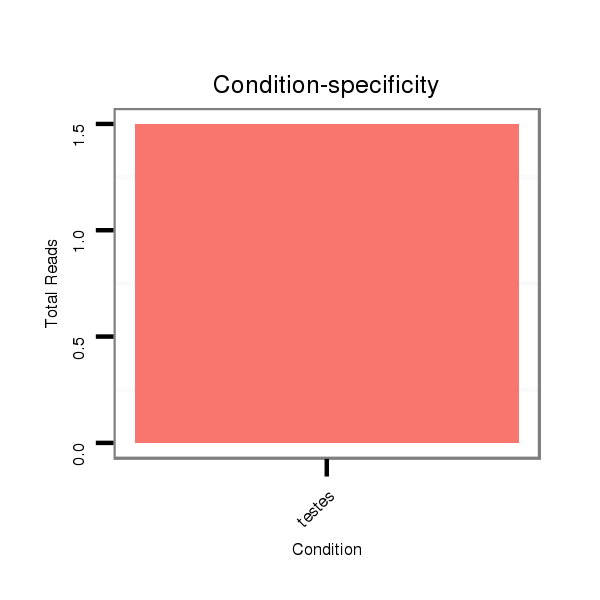
ID:eca-mir-9004 |
Coordinate:chr8:33967964-33968082 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| mature | star |
|
CTGTGATAGCAATTGCAGATTTTACTAGATACAGTTTCTTTCTTtTTTtTTTtTTTtTTTtTTTtGTGtGGttGtTTGGgtgtGTGTTtGgTgtGTGtgtGTTTTGTGtGGttGtTTGTgtgTGtGgTttgtgTGTGggtGTgTTggTgTtTTTTGTtTGTGGGtTGggTggtgtGgtTGGgTGtTGttTGGtGTtGGTgTGgtgggtGGtTggtttgg
*****************************************************************......................................................................................******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ................................................................................ACAGTGTTAGCTCAGTGACAGT..................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................ATTGTCACTGAGCTAACACTGT................................................................................... | 22 | 0 | 3 | 0.33 | 1 | 1 |
| ...............................................................TAGTGAGGAAGATTGGCAC......................................................................................................................................... | 19 | 0 | 6 | 0.17 | 1 | 1 |
| ..................................................................................................................................................................................TGGCTGATGAATGGAGTAGGTCTGCACC............. | 28 | 0 | 14 | 0.14 | 2 | 2 |
| .........................................................................................................................................................................................TGAATGGAGTAGGTCTGCACCCAGGAT....... | 27 | 0 | 13 | 0.08 | 1 | 1 |
| ......................................................................................................................................................................................TGATGAACGGAGTAGGTCTGCACCCAAGATA...... | 31 | 3 | 15 | 0.07 | 1 | 1 |
| ...................................................................................................................................................................................GGCTGATGAATGGAGTAGGTCTGCACCTATA......... | 31 | 3 | 20 | 0.05 | 1 | 1 |
| .........................................................................................................................................................................................TGAATGGAGTAGGTCTGCACC............. | 21 | 0 | 20 | 0.05 | 1 | 1 |
|
GACACTATCGTTAACGTCTAAAATGATCTATGTCAAAGAAAGAAtAAAtAAAtAAAtAAAtAAAtCACtCCttCtAACCgtgtCACAAtCgAgtCACtgtCAAAACACtCCttCtAACAgtgACtCgAttgtgACACggtCAgAAggAgAtAAAACAtACACCCtACggAggtgtCgtACCgACtACttACCtCAtCCAgACgtgggtCCtAggtttgg
********************************************************************......................................................................................***************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACATACAC......................................................... | 31 | 0 | 20 | 1.20 | 24 | 24 |
| ....................................................................................................................................GACACGGTCAGAAGGAGATAAAACATACAC......................................................... | 30 | 0 | 20 | 0.95 | 19 | 19 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACATA............................................................ | 28 | 0 | 20 | 0.75 | 15 | 15 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACATACA.......................................................... | 30 | 0 | 20 | 0.70 | 14 | 14 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACAT............................................................. | 27 | 0 | 20 | 0.40 | 8 | 8 |
| .................................................................................................................................TATGACACTGTCAGAAGGAGATAAAACATA............................................................ | 30 | 2 | 9 | 0.11 | 1 | 1 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACATACACC........................................................ | 32 | 0 | 20 | 0.05 | 1 | 1 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAAACATAC........................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 |
| ........................................................................................................ACACTCCTTCTAACAGTGACTCGATTGT....................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGATAAA................................................................. | 23 | 0 | 20 | 0.05 | 1 | 1 |
| ...................................................................................................................................TGACACGGTCAGAAGGAGAT.................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 |
| ...............................................................................................................................................................................................CTCATCCAGACGTGGGTCCTAGGTTT.. | 26 | 0 | 20 | 0.05 | 1 | 1 |
Generated: 09/01/2015 at 12:10 PM