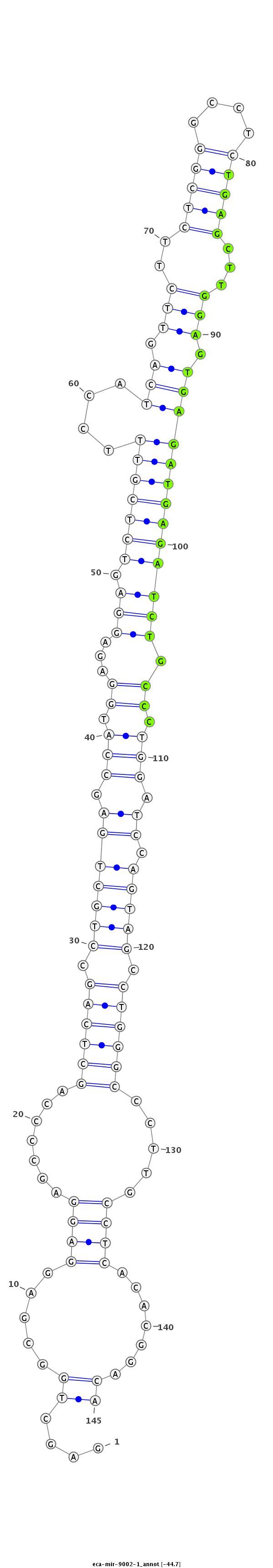
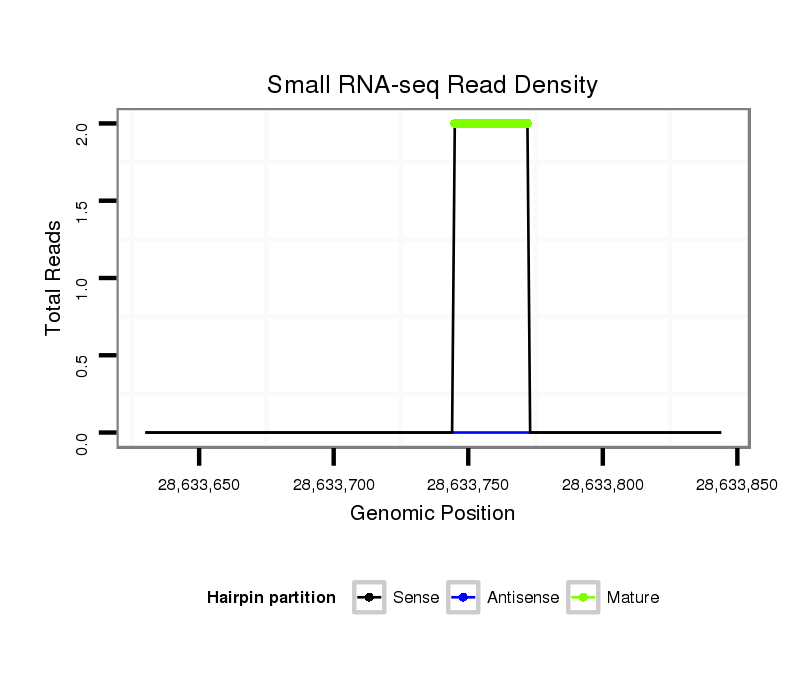
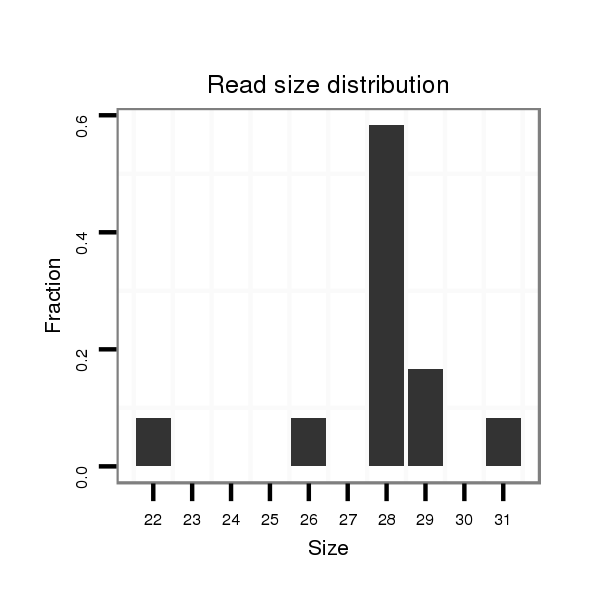
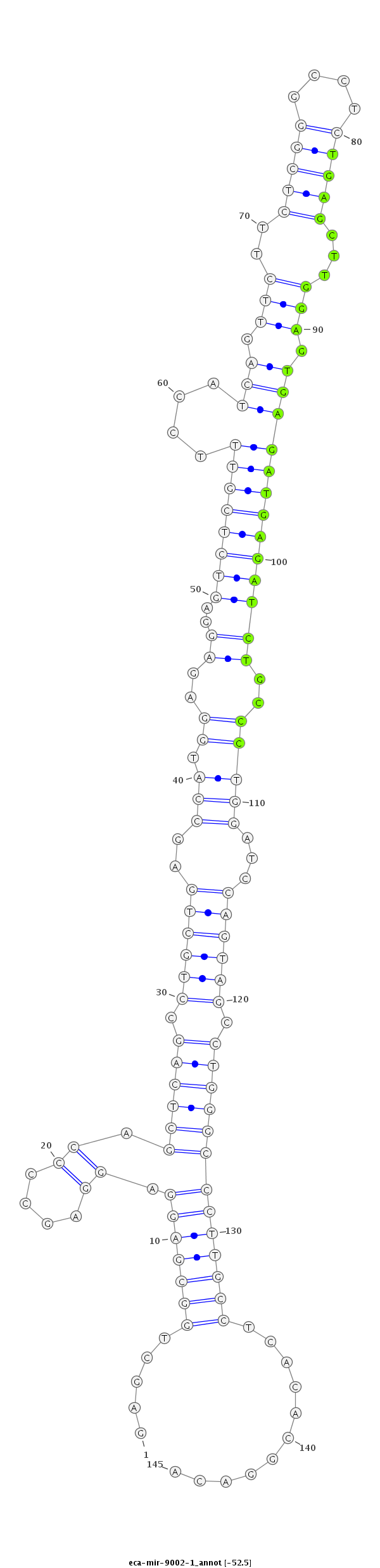
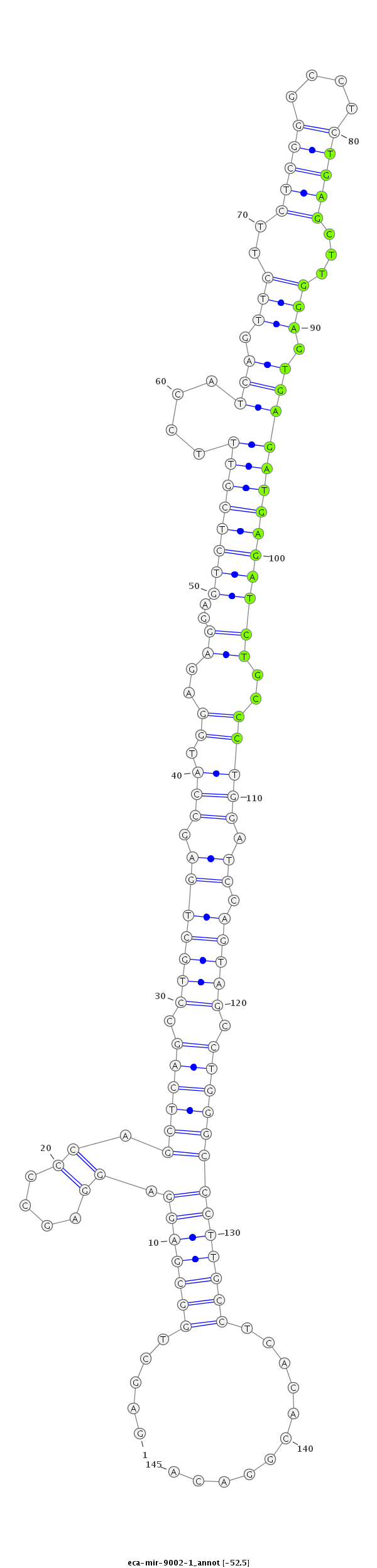
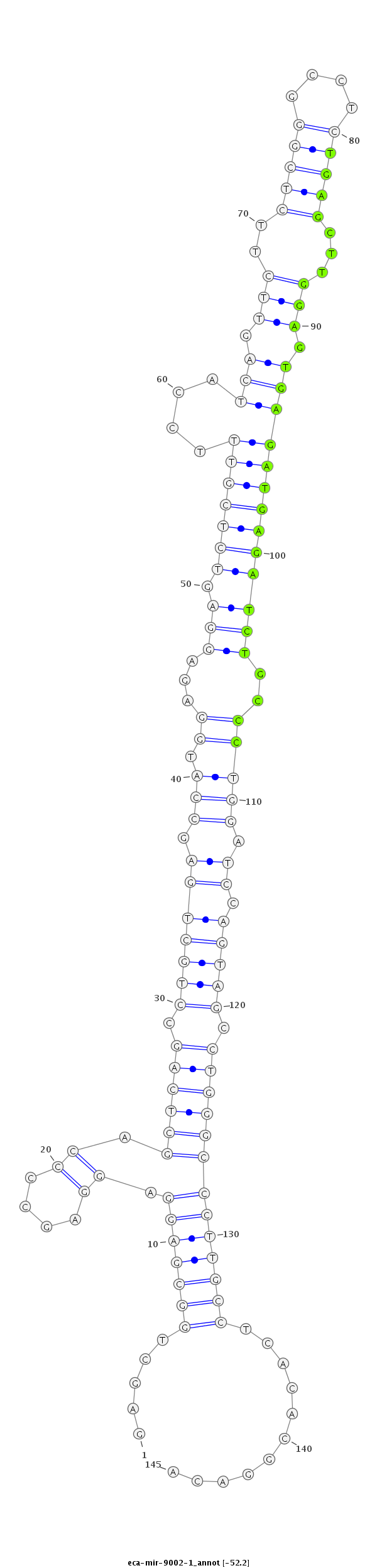
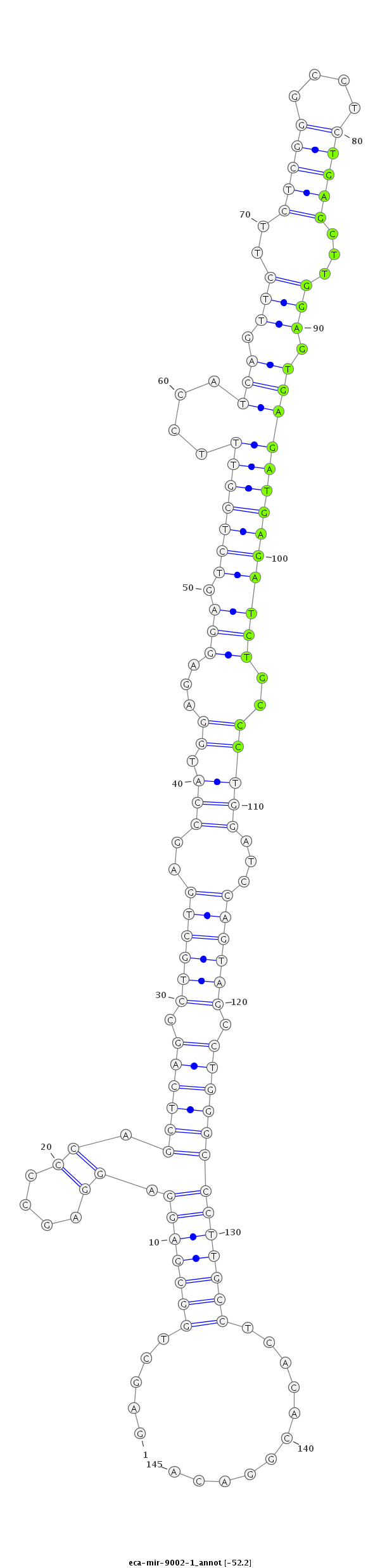
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGCCC........................................................................ |
28 |
0 |
3 |
2.33 |
7 |
7 |
| .........................................................................................................................................................................TCACACGGACACGAGAAGGAAACAAATG.................. |
28 |
1 |
2 |
1.00 |
2 |
2 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGAA......................................................................... |
27 |
2 |
3 |
0.67 |
2 |
2 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGCCT........................................................................ |
28 |
1 |
3 |
0.67 |
2 |
2 |
| ..........................................................................................................................................................................CACACGGACACGAGAAGGAAACAAATGCC................ |
29 |
1 |
2 |
0.50 |
1 |
1 |
| ........................................................................................................................................................TAGCCTGGGCCCTTGCCTCACACGGACACG................................. |
30 |
0 |
2 |
0.50 |
1 |
1 |
| ...........................................................................................TTCCATCAGTTCTTCTCGGGCCTCTGT................................................................................................. |
27 |
1 |
3 |
0.33 |
1 |
1 |
| ...........................................................................................TTCCATCAGTTCTTCTCGGGCCTCTGAGCTTT............................................................................................ |
32 |
1 |
3 |
0.33 |
1 |
1 |
| ..........................................................................................TTTCCATCAGTTCTTCTCGGGCCTCTGAG................................................................................................ |
29 |
0 |
3 |
0.33 |
1 |
1 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGCT......................................................................... |
27 |
1 |
3 |
0.33 |
1 |
1 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGCCCT....................................................................... |
29 |
0 |
3 |
0.33 |
1 |
1 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCCGCCT........................................................................ |
28 |
2 |
3 |
0.33 |
1 |
1 |
| ...........................................................................................TTCCATCAGTTCTTCTCGGGCC...................................................................................................... |
22 |
0 |
3 |
0.33 |
1 |
1 |
| ................................................................................................TCAGTTCTTCTCGGGCCTCTGAGCTTGGAGT........................................................................................ |
31 |
0 |
3 |
0.33 |
1 |
1 |
| ...................................................................................................................TGAGCTTGGAGTGAGATGAGATCTGC.......................................................................... |
26 |
0 |
3 |
0.33 |
1 |
1 |