
ID:eca-mir-8990 |
Coordinate:chr7:98037125-98037269 - |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
GTtggGgTGGGtGGGTggTTTGtggggtGgTGGTGtGgttgggTGtGgtGGTGtGggGGGTGGgTGGGggttGGGGgTGGGTGgTGGGtGTGTGgTTTgTGtgTGGGTGGgtgGTGGggTGGGGgTGTGGTGTTGtgTTTGGGtGGGggggGGtttggGTgGTggtGGGTGgtGtTttGggTTGGTGgGgtGGgGGTGTGgtGGGgtgTTtGGGtGggTGttGGGGtGGggGgtGTGtGGTGGTg
********************************************************************************..................................................................................................******************************************************************* |
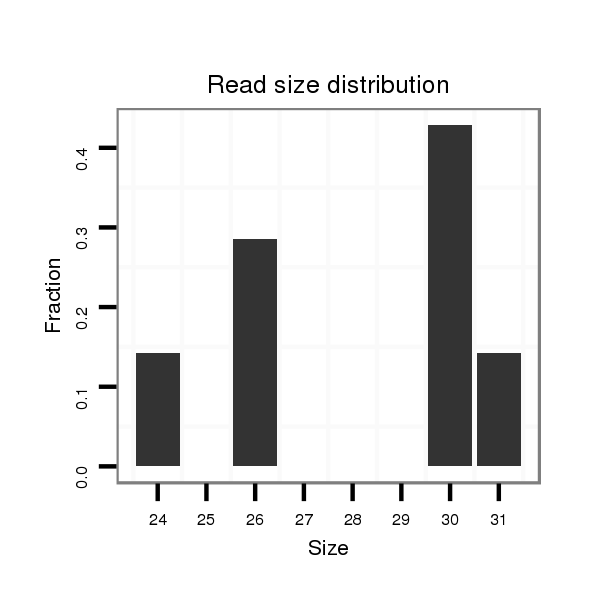
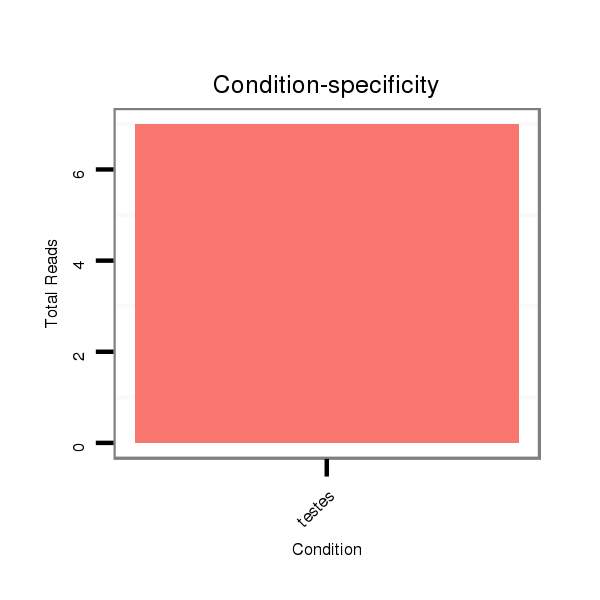
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .....................................................................................................................................TGACTTTGGGAGGGCCCCGGAAACCGTCGT.................................................................................. | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ..................TTTGACCCCAGCTGGTGAGCAACCCT......................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ............................................................................................TGCTTTCTGACTGGGTGGCACGTGGCCTT............................................................................................................................ | 29 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGTTGACTTTGGGAGGGCCCCGGAAACCGTC.................................................................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ..................TTTGACCCCAGCTGGTGAGCAACCCTGAGCTTT.................................................................................................................................................................................................. | 33 | 3 | 1 | 1.00 | 1 | 1 |
| ......................................................................................................................................................................................................TGCAGGGCACTTAGGGAGCCTGAAGGGGAA................. | 30 | 1 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................TTTCTGACTGGGTGGCACGTGGCCTG............................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................................................TGGTGTTGACTTTGGGAGGGCCCCGGAT.......................................................................................... | 28 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................................................................................AAACCGTCGTCCAGGGTGCAGATAAGCCTT.............................................................. | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ............................................................................................TGCTTTCTGACTGGGTGGCACGTGGCCTGTAA......................................................................................................................... | 32 | 3 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................TTTCTGACTGGGTGGCACGTGGCCTGGGGC........................................................................................................................ | 30 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................TGGGAGTGTGCTTTCTGACTGGGTGG....................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ..................TTTGACCCCAGCTGGTGAGCAACCCTGA....................................................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................TGTTGACTTTGGGAGGGCCCCGGA........................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
|
CAtggCgACCCtCCCAggAAACtggggtCgACCACtCgttgggACtCgtCCACtCggCCCACCgACCCggttCCCCgACCCACgACCCtCACACgAAAgACtgACCCACCgtgCACCggACCCCgACACCACAACtgAAACCCtCCCggggCCtttggCAgCAggtCCCACgtCtAttCggAACCACgCgtCCgCCACACgtCCCgtgAAtCCCtCggACttCCCCtCCggCgtCACtCCACCAg
*******************************************************************..................................................................................................******************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 12:07 PM