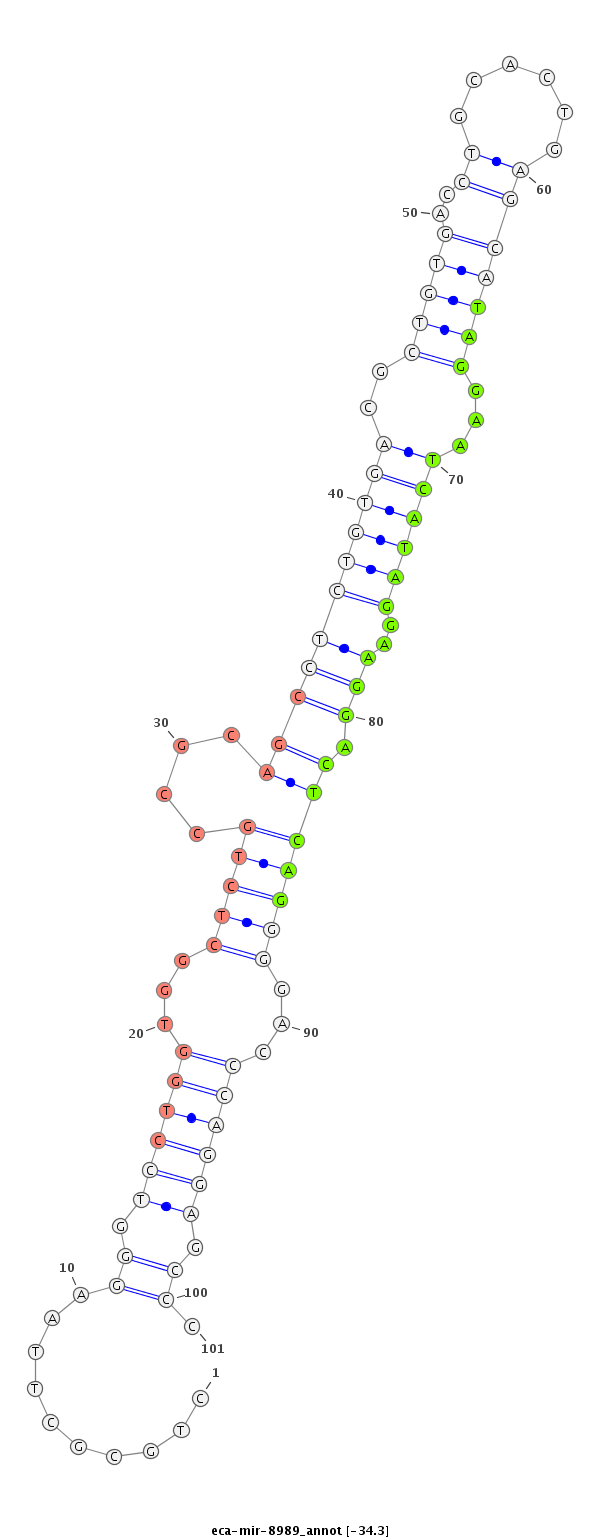
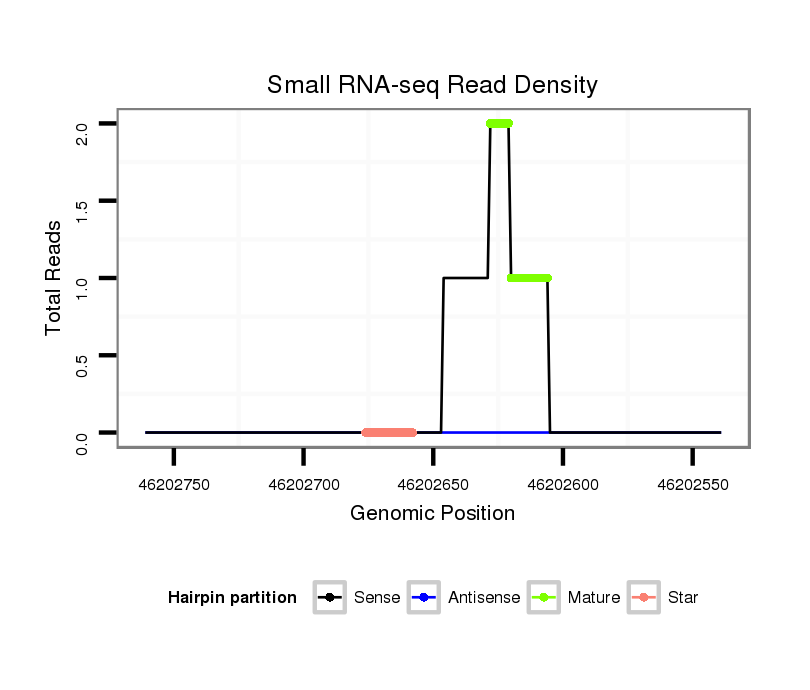
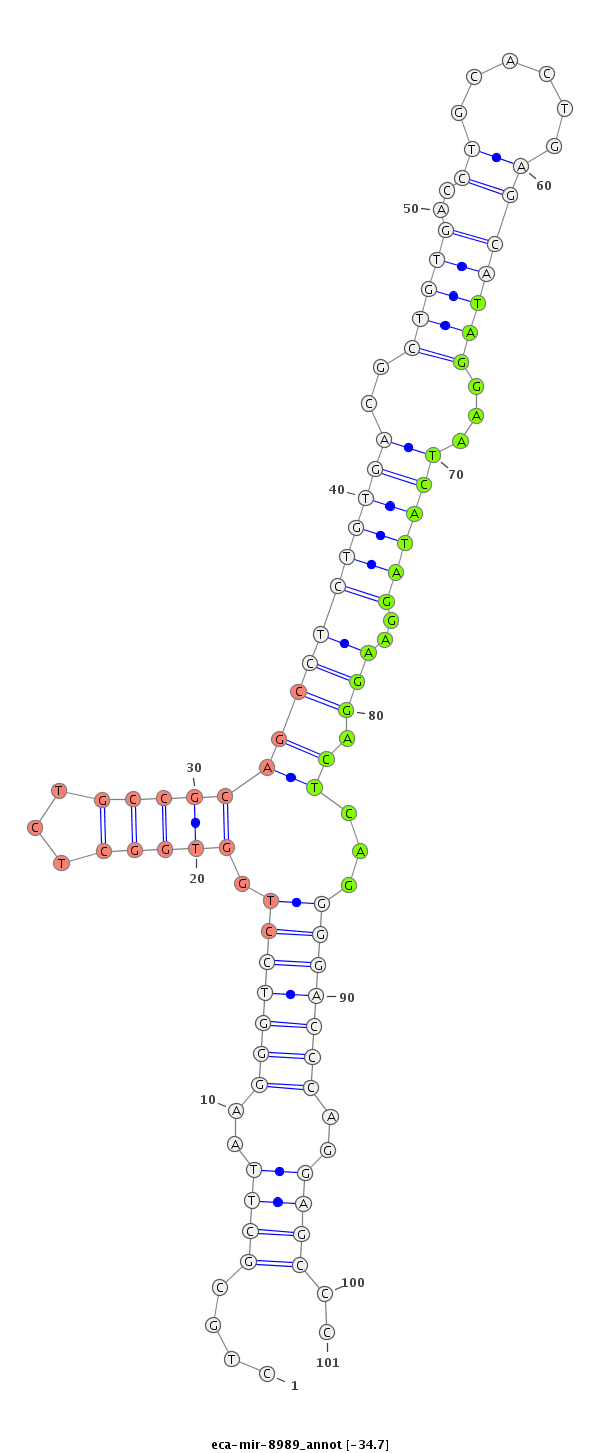
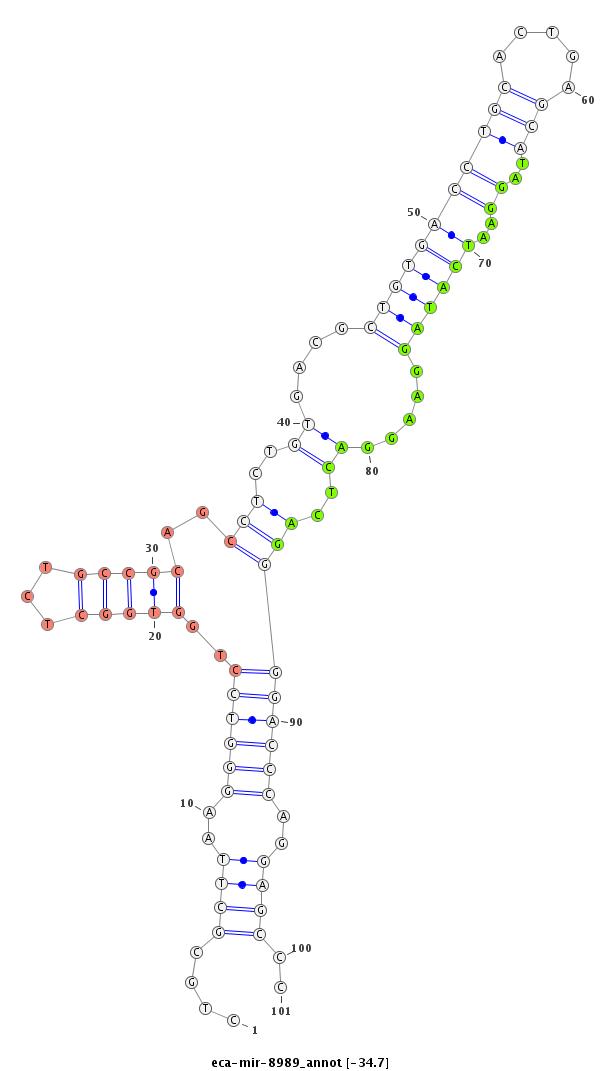
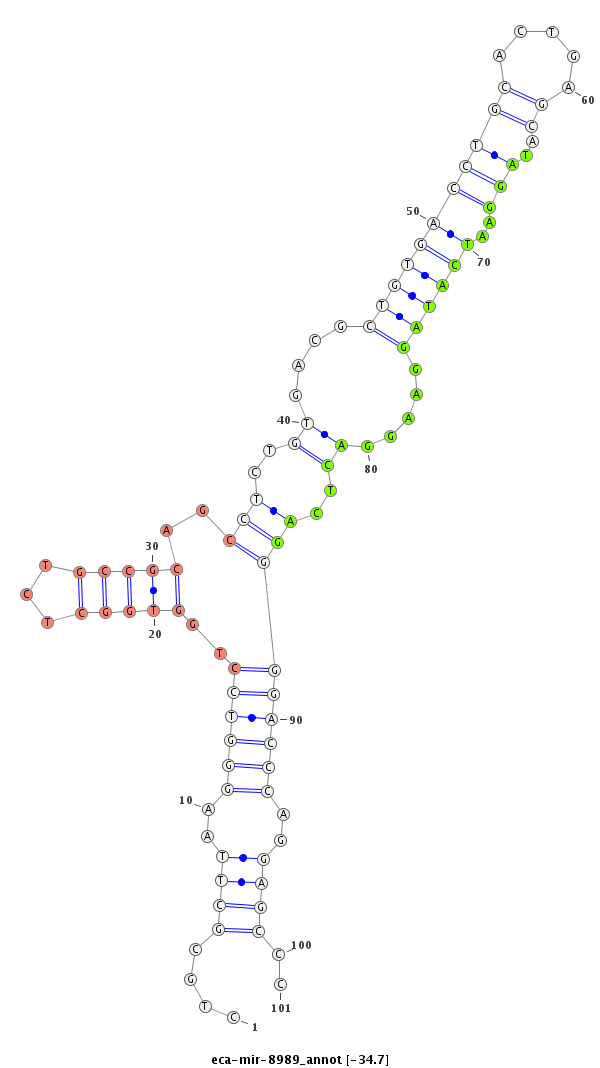
| .....................................................................................................................................TAGGAATCATAGGAAGGACTCAG................................................................... |
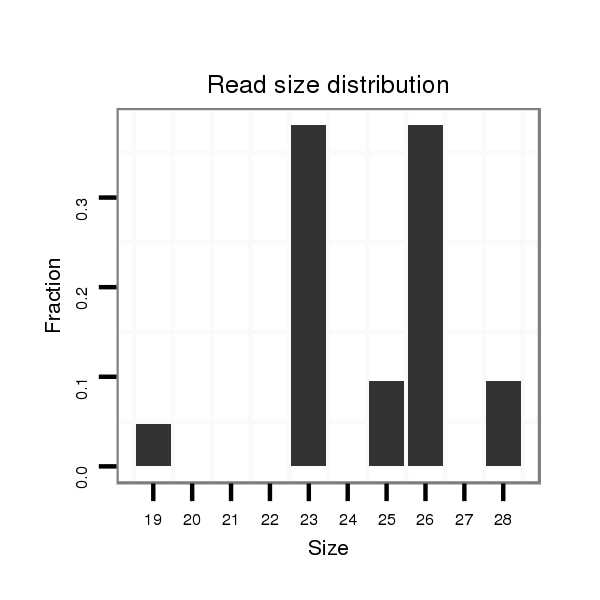
23 |
0 |
1 |
1.00 |
1 |
1 |
| ...................................................................................................................TGTGACCTGCACTGAGCATAGGAATC.................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAGTCC.................................................... |
29 |
1 |
7 |
0.43 |
3 |
3 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAA....................................................... |
26 |
1 |
9 |
0.33 |
3 |
3 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGA........................................................ |
25 |
0 |
8 |
0.25 |
2 |
2 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAGCC..................................................... |
28 |
0 |
4 |
0.25 |
1 |
1 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAGT...................................................... |
27 |
1 |
8 |
0.25 |
2 |
2 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGATT...................................................... |
27 |
2 |
9 |
0.22 |
2 |
2 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAT....................................................... |
26 |
1 |
8 |
0.13 |
1 |
1 |
| .....................................................................................CTGGTGGCTCTGCCGCAGC....................................................................................................................... |
19 |
0 |
8 |
0.13 |
1 |
1 |
| ..............................................................................................................................................TAGGAACGAGTCAGGGGACCCAGGA........................................................ |
25 |
2 |
9 |
0.11 |
1 |
1 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAAT...................................................... |
27 |
2 |
10 |
0.10 |
1 |
1 |
| ..............................................................................................................................................TAGGAAGGACTCAGGGGACCCAGGAGTTT.................................................... |
29 |
3 |
11 |
0.09 |
1 |
1 |
| ..............................................................................................................................................TAGGAAGGACTCCGGGGACCCAGGATT...................................................... |
27 |
3 |
20 |
0.05 |
1 |
1 |