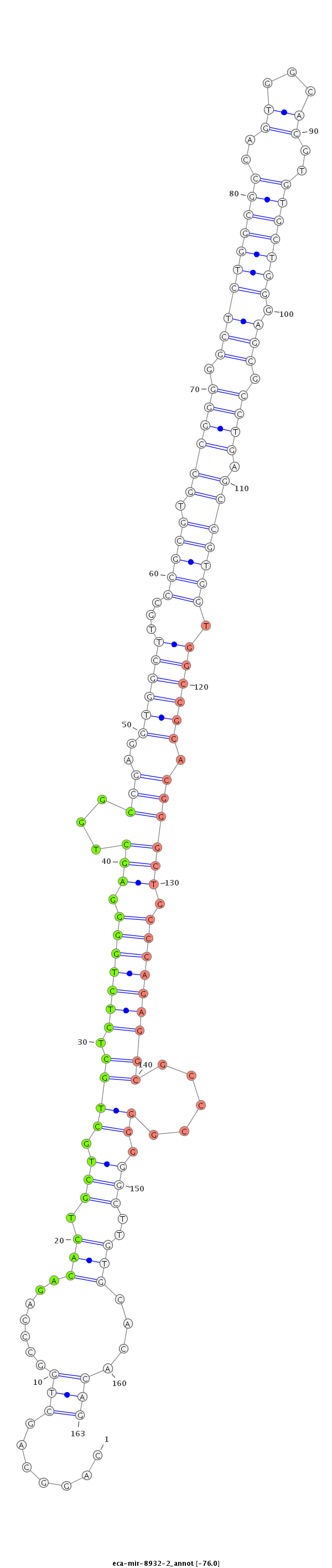
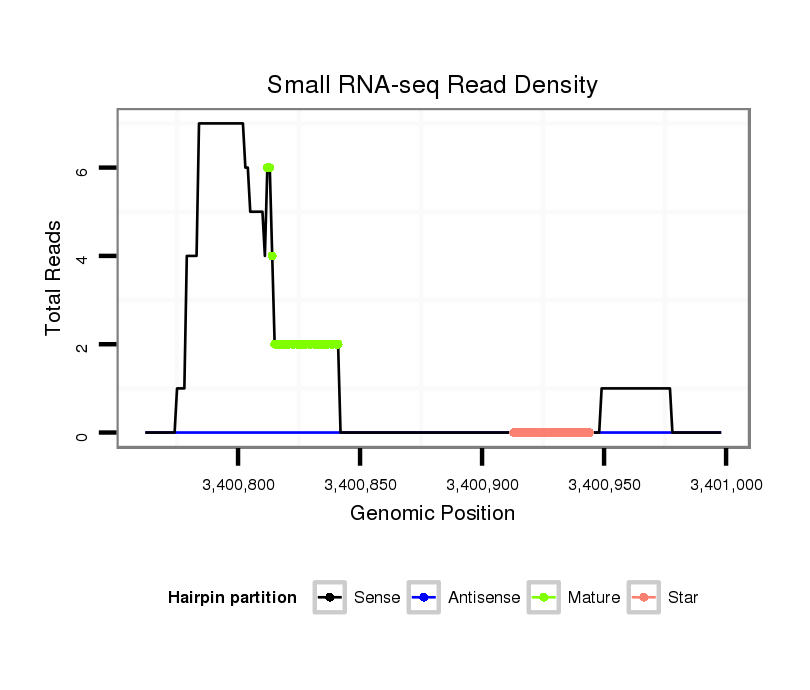
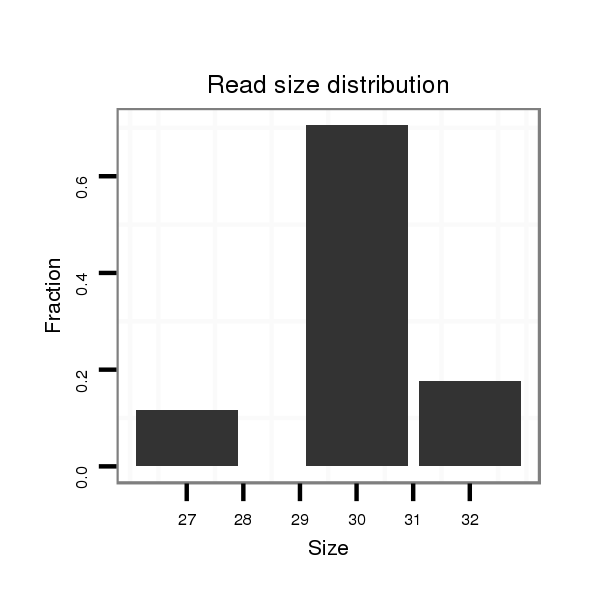
| ......................TGGACGTGTGGACCAGGCAGCTGGCCCAGA......................................................................................................................................................................................... |
30 |
0 |
1 |
2.00 |
2 |
2 |
| .................TAGCATGGACGTGTGGACCAGGCAGCTGGCCCCT.......................................................................................................................................................................................... |
34 |
2 |
1 |
2.00 |
2 |
2 |
| ..................................................GACACTGCTGCTGCTCTCTGGGGAGCTGGC............................................................................................................................................................. |
30 |
0 |
1 |
2.00 |
2 |
2 |
| .................TAGCATGGACGTGTGGACCAGGCAGCTGGCCT............................................................................................................................................................................................ |
32 |
1 |
1 |
1.00 |
1 |
1 |
| ......................TGGACGTGTGGACCAGGCAGCTGGCCCAGACTTC..................................................................................................................................................................................... |
34 |
3 |
1 |
1.00 |
1 |
1 |
| ............GGCCATTGCCTGGACGTGTGGACCAGGC..................................................................................................................................................................................................... |
28 |
2 |
1 |
1.00 |
1 |
1 |
| ...........TGGCCATAGCATGGACGTGTGGACCAGGCT.................................................................................................................................................................................................... |
30 |
1 |
1 |
1.00 |
1 |
1 |
| .................TAGCATGGACGTGTGGACCAGGCAGCTGGCACA........................................................................................................................................................................................... |
33 |
1 |
1 |
1.00 |
1 |
1 |
| ......................TGGACGTGTGGACCAGGCAGCTGGCCCAGAC........................................................................................................................................................................................ |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .................TAGCATGGACGTGTGGACCAGGCAGCTGGCCCAGAC........................................................................................................................................................................................ |
36 |
0 |
1 |
1.00 |
1 |
1 |
| .................TAGCATGGACGTGTGGACCAGGCAGCTGGCCC............................................................................................................................................................................................ |
32 |
0 |
1 |
1.00 |
1 |
1 |
| .................TAGCATGGACGTGTGGACCAGGCAGC.................................................................................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................................................................................................................TGTGCACACAGCGGGCCGGCTCTTGGCCC..................... |
29 |
0 |
2 |
1.00 |
2 |
2 |
| .............GCCATAGCATGGACGTGTGGACCAGGCA.................................................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................................................................................................................TGCGCTGGGAGCGCCTGAGCCATGGTGG................................................................................... |
28 |
2 |
3 |
0.67 |
2 |
2 |
| ...........................................................................................................................................................................................TGTGCACACAGCGCGCCAGCTCTTGGCCC..................... |
29 |
2 |
2 |
0.50 |
1 |
1 |
| ..........................................................................................................................................................................................TTGTGCACACAGCGGGCCGGCTCTTGGCCCT.................... |
31 |
1 |
2 |
0.50 |
1 |
1 |
| ..........................................................................................................................................................................................TTGTGCACACAGCGGGCCGGCCCTTGGCTT..................... |
30 |
3 |
2 |
0.50 |
1 |
1 |
| .......................................................................................................................................................TGGCCGCACGGGCTGCCCAGAGGCGCCCGGGG...................................................... |
32 |
0 |
2 |
0.50 |
1 |
1 |
| .......................................................................................................................TGGCACGTGCGCTGGGAGCGCCTGAGC........................................................................................... |
27 |
1 |
2 |
0.50 |
1 |
1 |
| ...........................................................................................................................................................................................TGTGCACACAGCGGGCCGGCTCTTGG........................ |
26 |
0 |
2 |
0.50 |
1 |
1 |
| ..................................................................................................TGCCGGGGGCTCTGGCGCCAGTGGCAC................................................................................................................ |
27 |
0 |
3 |
0.33 |
1 |
1 |