ID:eca-mir-8913 |
Coordinate:chr20:39845361-39845485 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
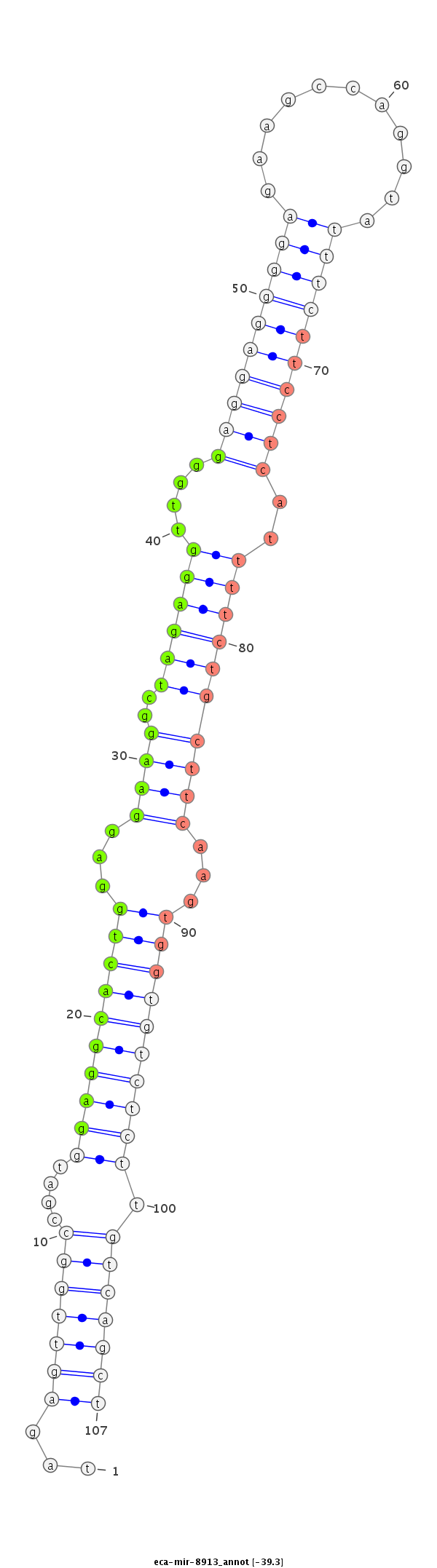
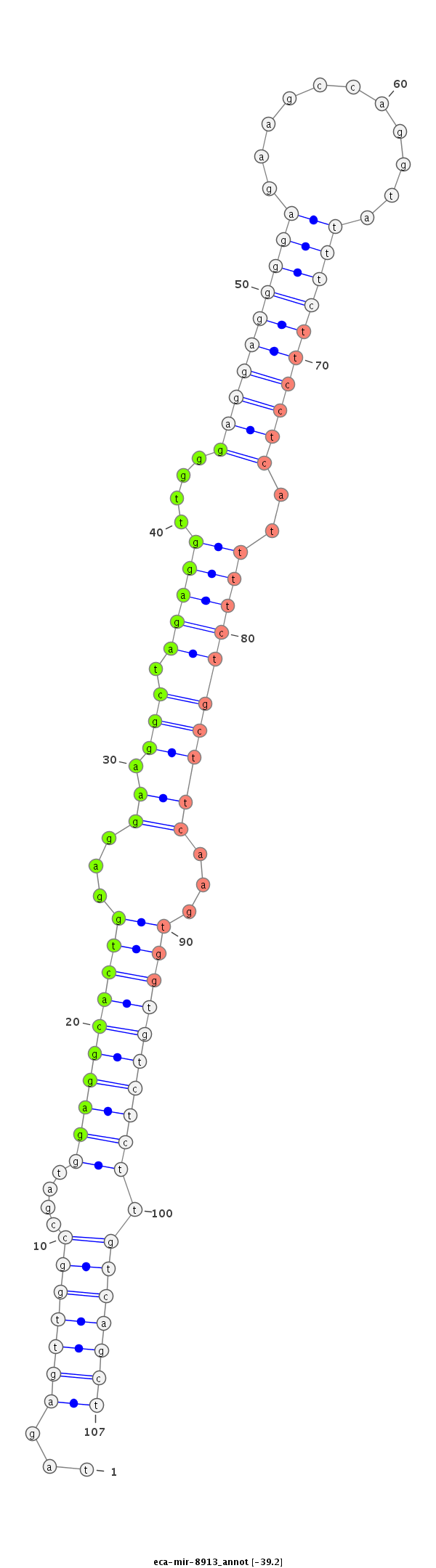
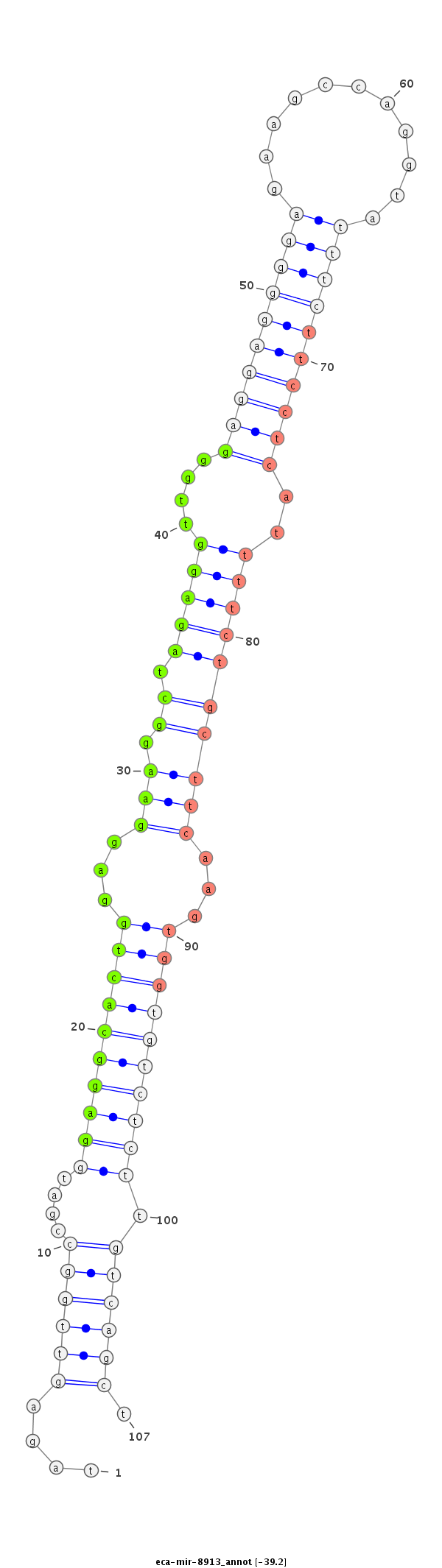
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -39.3 | -39.2 | -39.2 |
 |
 |
 |
|
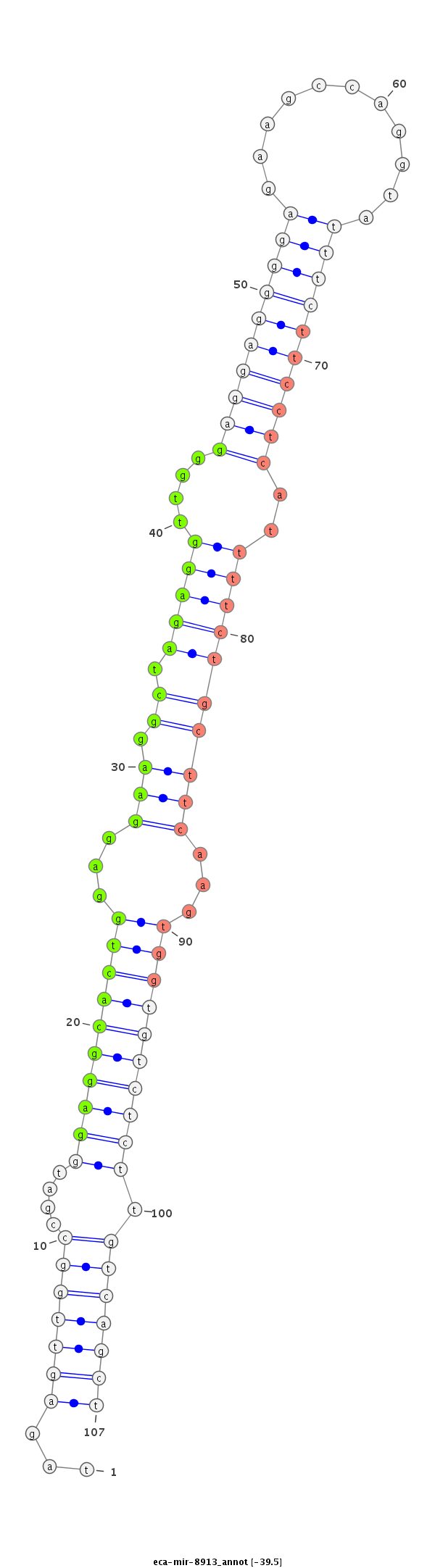
ttcccagactcccttgcctgttcccaggctcctttgcccttgggcttccaggtagagttggccgatggaggcactggaggaaggctagaggttgggaggaggggagaagccaggtatttcttcctcattttctgcttcaagtggtgtctcttgtcagctcctgagcctcctctgtgatcccagctccctgggttctagctcctgccggatggatttggattttca
****************************************************...(((((((....((((((((((...(((.((.(((((....((((((((((...........))))))))))..))))))))))...)))))))))).)))))))****************************************************************** |
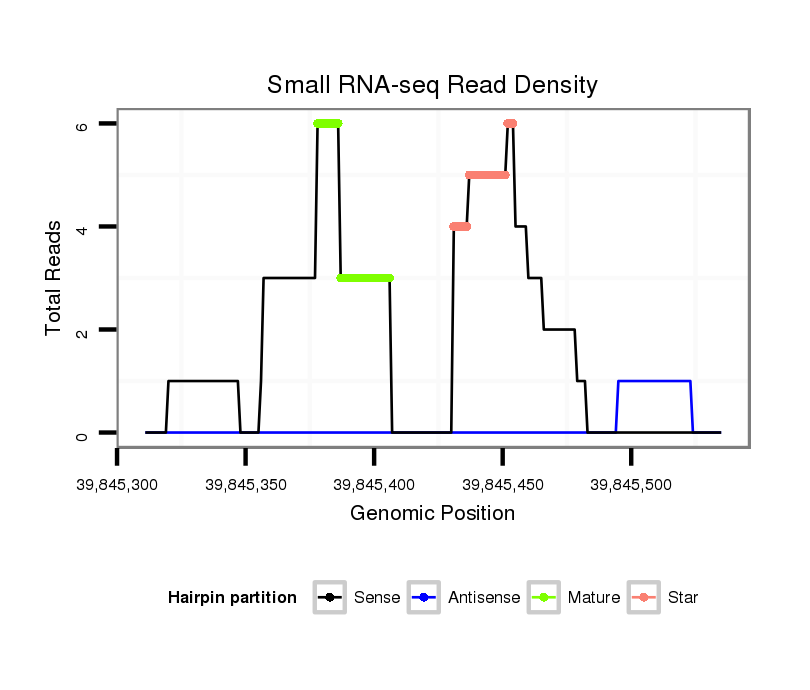
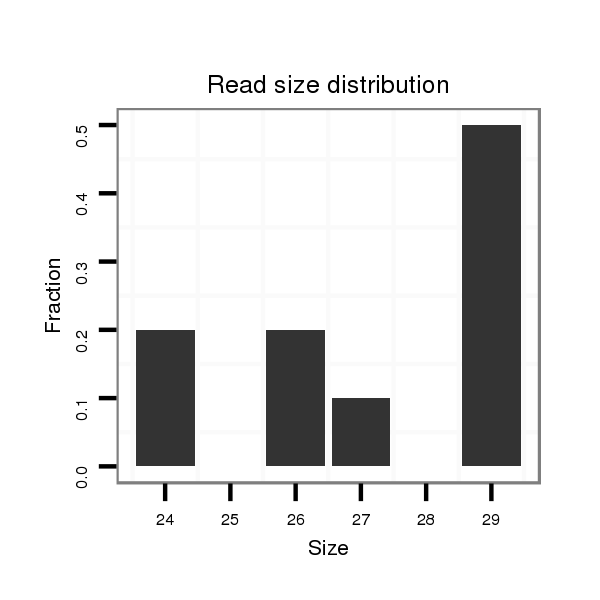
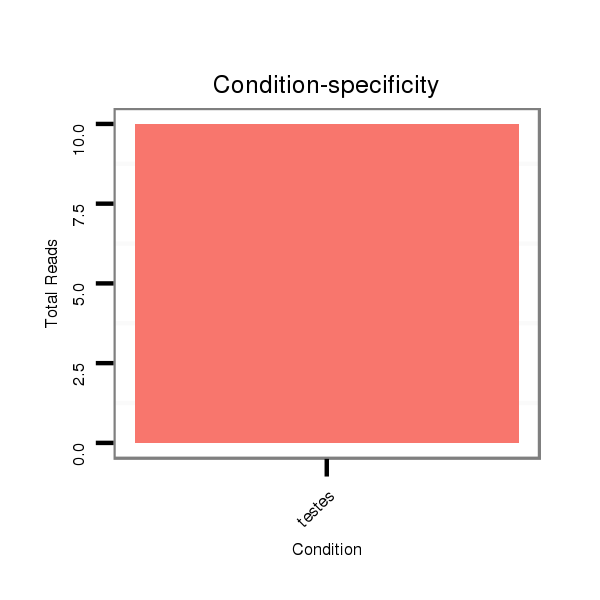
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...................................................................GAGGCACTGGAGGAAGGCTAGAGGTTGGG................................................................................................................................. | 29 | 0 | 1 | 3.00 | 3 | 3 |
| ........................................................................................................................TTCCTCATTTTCTGCTTCAAGTGG................................................................................. | 24 | 0 | 1 | 2.00 | 2 | 2 |
| ..............................................TCCAGGTAGAGTTGGCCGATGGAGGCACTG..................................................................................................................................................... | 30 | 0 | 1 | 2.00 | 2 | 2 |
| .........TCCCTTGCCTGTTCCCAGGCTCCTTTGC............................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 |
| ..............................................................................................................................ATTTTCTGCTTCAAGTGGTGTCTCTTGTC...................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................................................TGGTGTCTCTTGTCAGCTCCTGAGCCT......................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ........................................................................................................................TTCCTCATTTTCTGCTTCAAGTGGTG............................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................TTCCAGGTAGAGTTGGCCGATGGAGGCACTG..................................................................................................................................................... | 31 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................................................................TCTTGTCAGCTCCTGAGCCTCCTAATT.................................................. | 27 | 3 | 1 | 1.00 | 1 | 1 |
| ........................................................................................................................TTCCTCATTTTCTGCTTCAAGTGGTGTCT............................................................................ | 29 | 0 | 1 | 1.00 | 1 | 1 |
| ........................................TGGGCTTCCAGGTAGAGTTGGCCGATGGAT........................................................................................................................................................... | 30 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................................................TCTCTTGTCAGCTCCTGAGCCTCCTC..................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ....................................................................................................................................................TCTTGTCAGCTCCTGAGCCTCCTCTGA.................................................. | 27 | 1 | 1 | 1.00 | 1 | 1 |
|
ttGGGTgTGtGGGttgGGtgttGGGTggGtGGtttgGGGttgggGttGGTggtTgTgttggGGgTtggTggGTGtggTggTTggGtTgTggttgggTggTggggTgTTgGGTggtTtttGttGGtGTttttGtgGttGTTgtggtgtGtGttgtGTgGtGGtgTgGGtGGtGtgtgTtGGGTgGtGGGtgggttGtTgGtGGtgGGggTtggTtttggTttttGT
******************************************************************...(((((((....((((((((((...(((.((.(((((....((((((((((...........))))))))))..))))))))))...)))))))))).)))))))**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................AGGGACCCAAGATCGAGGACGGCCTACCT............ | 29 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:49 AM