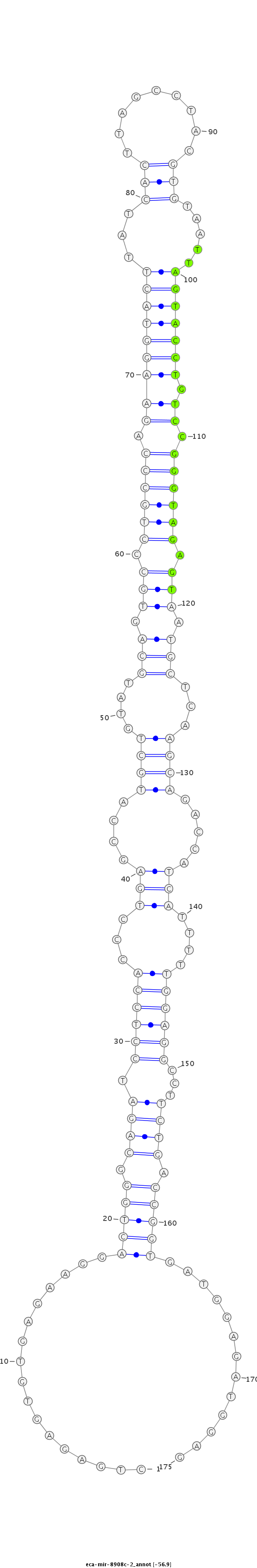
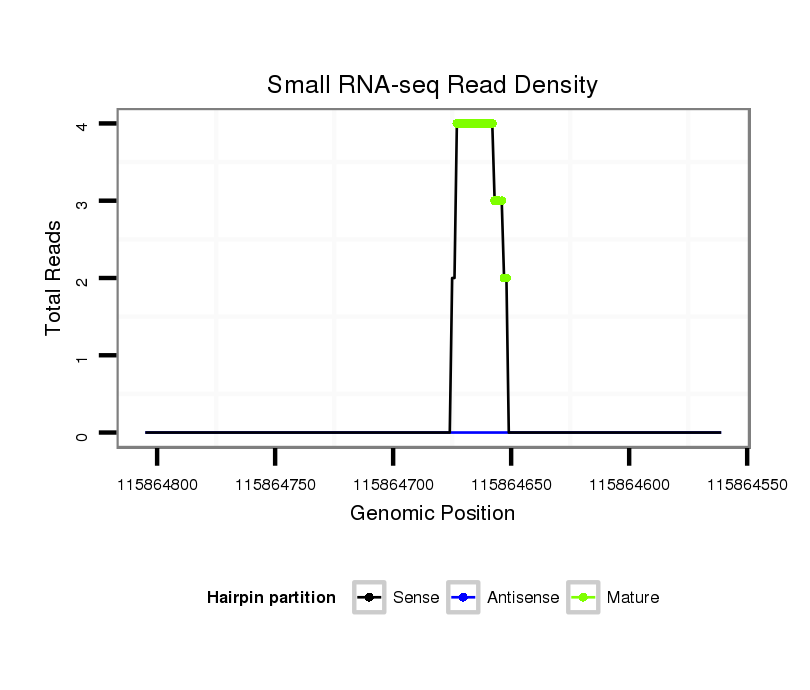
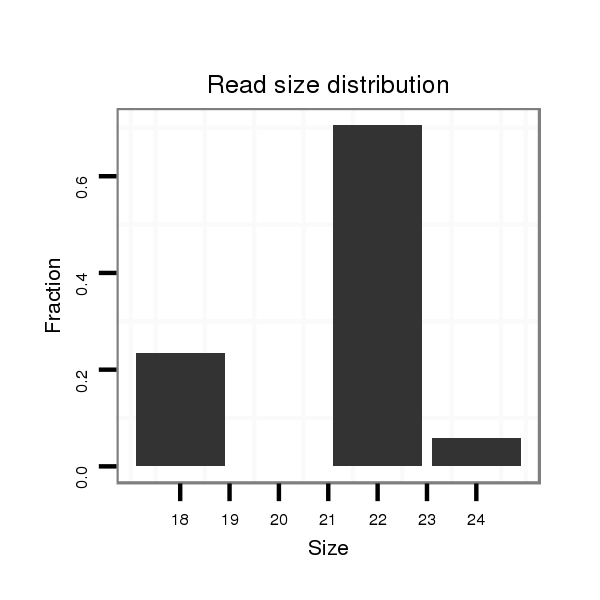
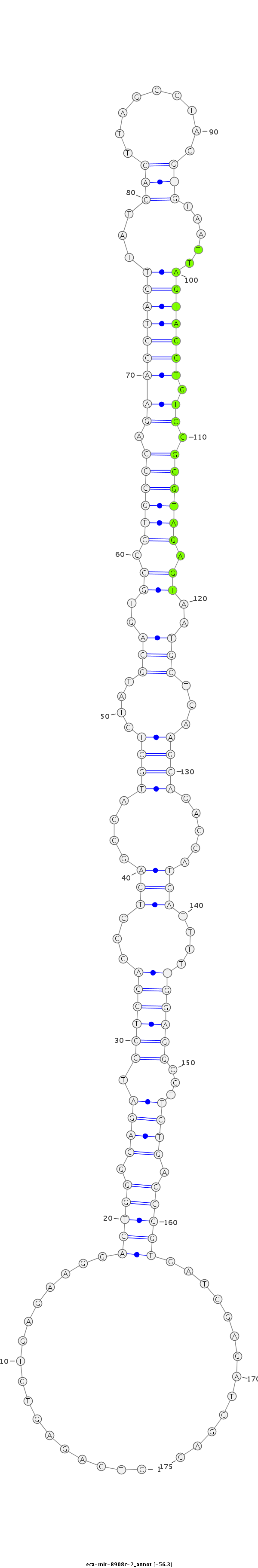
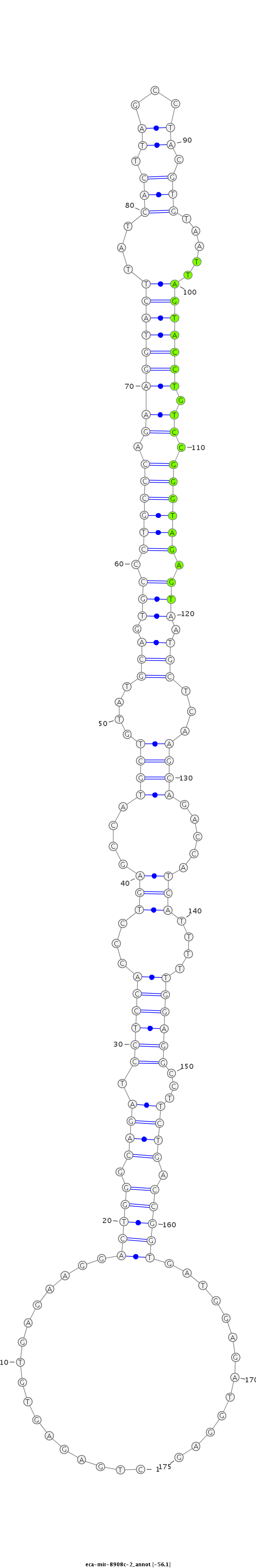
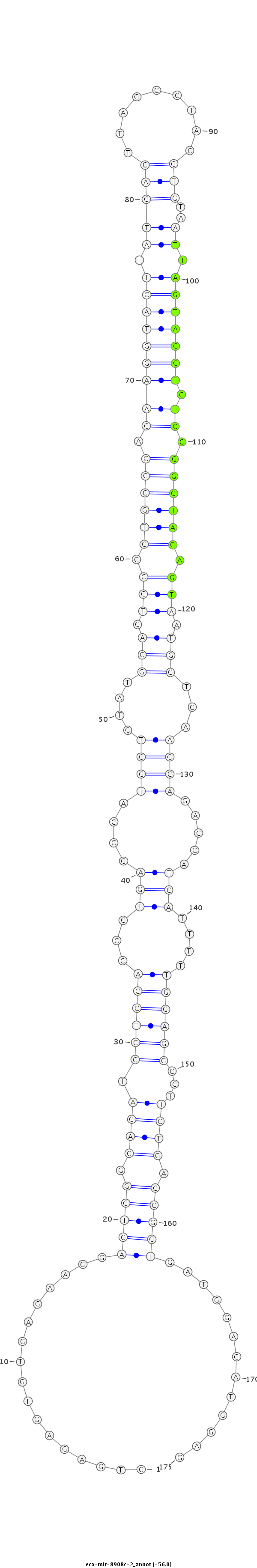
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAGT........................................................................................... |
22 |
0 |
3 |
2.33 |
7 |
7 |
| ..................................................................................................................................AATTAGTACCTGTCCGGGTAGA............................................................................................. |
22 |
0 |
3 |
1.67 |
5 |
5 |
| ..................................................................................................................................AATTAGTACCTGTCCGGG................................................................................................. |
18 |
0 |
3 |
1.33 |
4 |
4 |
| ..................................................................................................................................AATTAGTACCTGTCCGGGTAGAAT........................................................................................... |
24 |
1 |
3 |
1.00 |
3 |
3 |
| .....................................GAGAGTGTGAGAAGGACTGGGCAGATCC.................................................................................................................................................................................... |
28 |
0 |
2 |
0.50 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAAA........................................................................................... |
22 |
2 |
3 |
0.33 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAGTAT......................................................................................... |
24 |
1 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................................................AATTAGTACCTGTCCGGGTAGAT............................................................................................ |
23 |
1 |
3 |
0.33 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAGTT.......................................................................................... |
23 |
1 |
3 |
0.33 |
1 |
1 |
| .................................................................................................................................TAATTAGTACCTGTCCGGGTAAA............................................................................................. |
23 |
1 |
3 |
0.33 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAGAA.......................................................................................... |
23 |
1 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................................................AATTAGTACCTGTCCGGGTAGAATT.......................................................................................... |
25 |
2 |
3 |
0.33 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCCGGGTAGAA............................................................................................ |
21 |
1 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................................................AATTAGTACCTGTCCGGGTAGAGT........................................................................................... |
24 |
0 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................................................AATTAGTACCTGGCCGGGTAGA............................................................................................. |
22 |
1 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................................................CATTAGTACCTGTCCGGGTAGT............................................................................................. |
22 |
2 |
6 |
0.17 |
1 |
1 |
| ..................................................................................................................................CATTAGTACCTGTCCGGG................................................................................................. |
18 |
1 |
8 |
0.13 |
1 |
1 |
| ..................................................................................................................................AATTAGTACCTGTCGGGG................................................................................................. |
18 |
1 |
8 |
0.13 |
1 |
1 |
| ....................................................................................................................................TTAGTACCTGTCGGGGTAGAGT........................................................................................... |
22 |
1 |
11 |
0.09 |
1 |
1 |