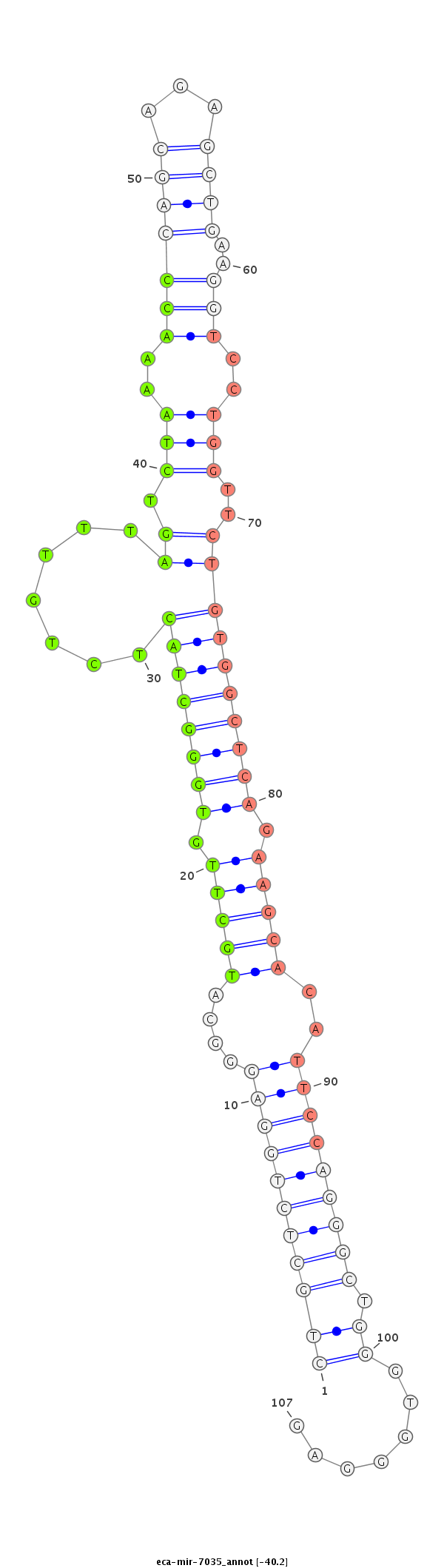
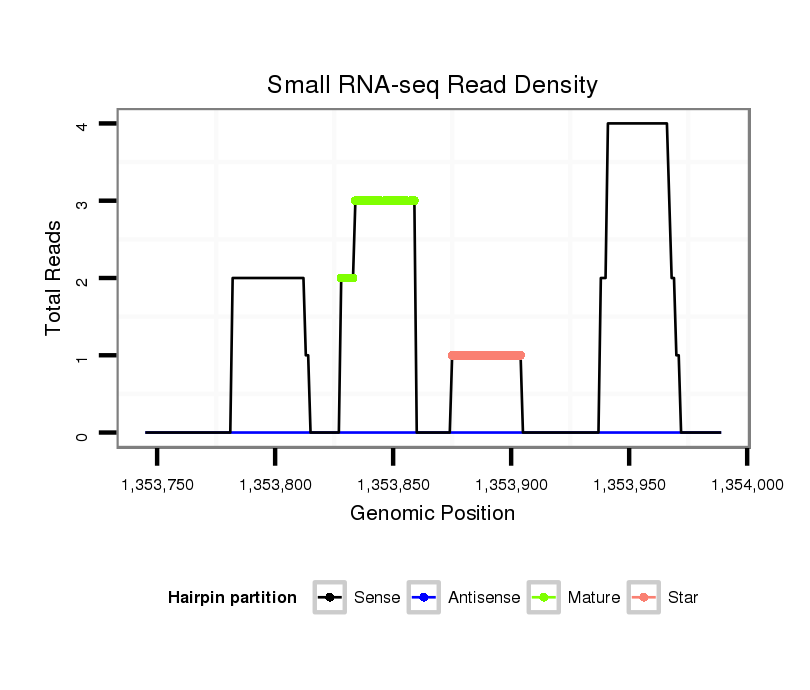
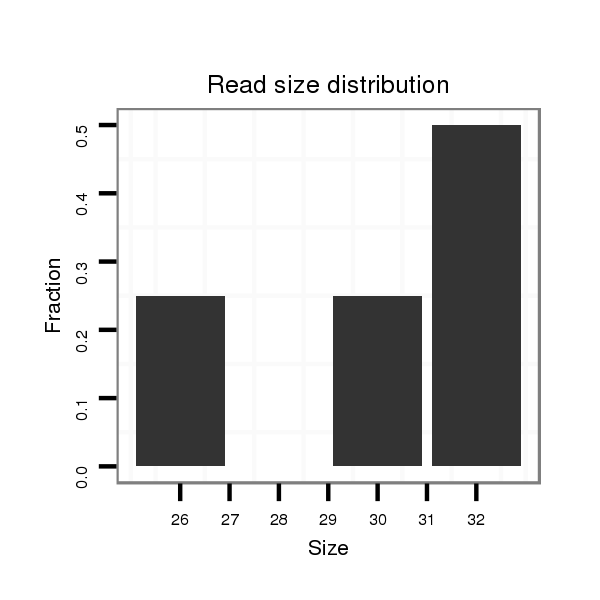
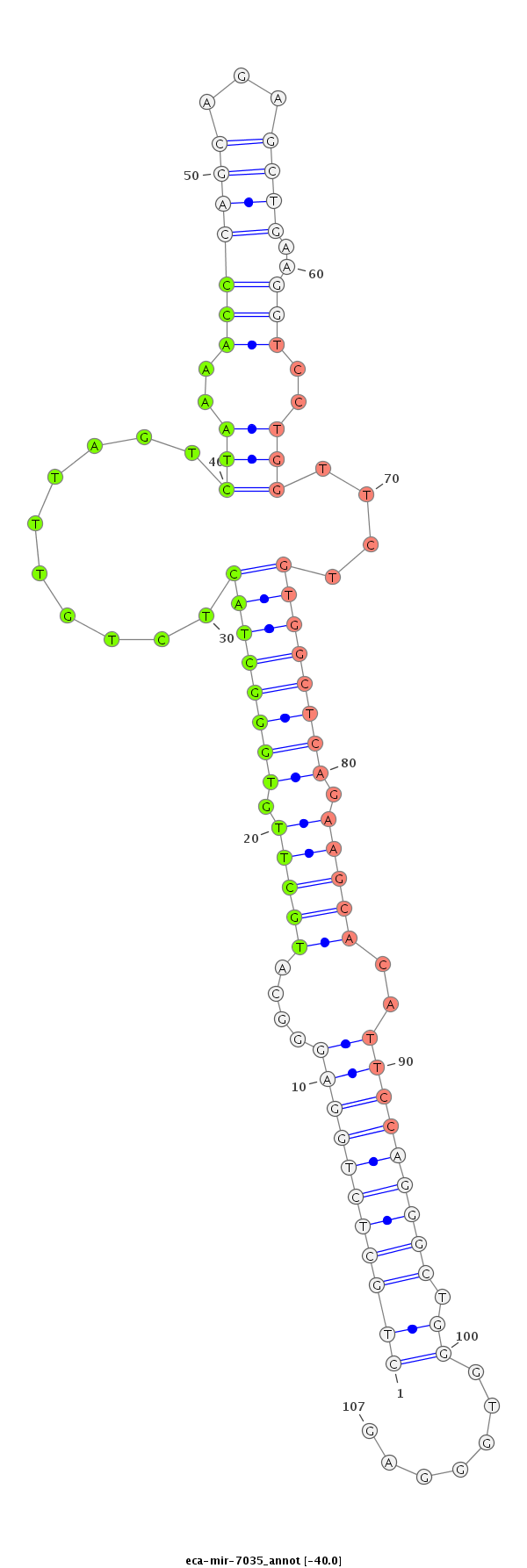
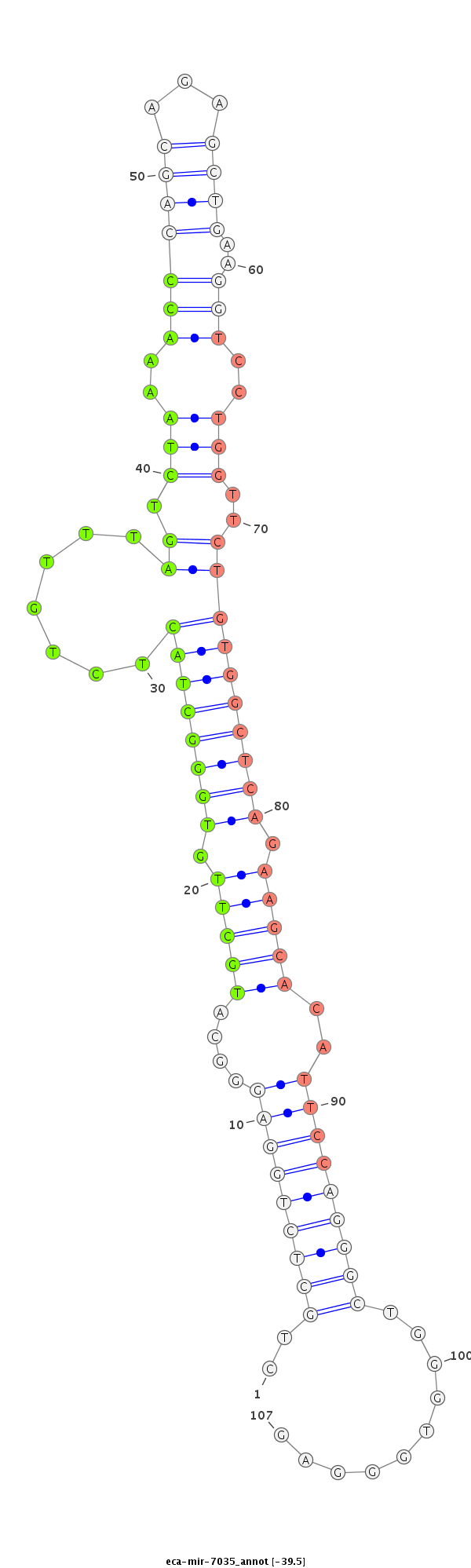
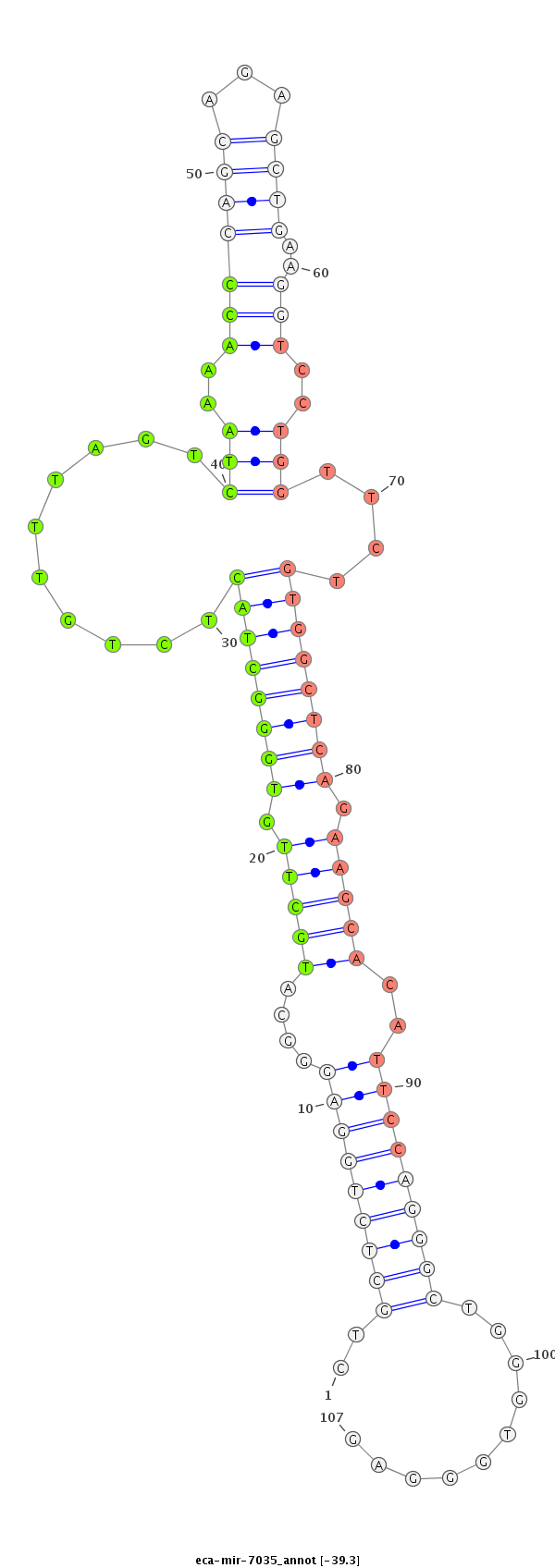
| ...................................................................................TGCTTGTGGGCTACTCTGTTTAGTCTAAAACC.................................................................................................................................. |
32 |
0 |
1 |
2.00 |
2 |
2 |
| ............................................................................................................................................................................................................TTAGAAGATGACAATAGCCCTTTGAGAACAGT......... |
32 |
1 |
1 |
2.00 |
2 |
2 |
| .....................................TCTGTAGTCTCTTTTGGGTCTTGCACTTCCCCT............................................................................................................................................................................... |
33 |
0 |
1 |
1.00 |
1 |
1 |
| ............................................................................................................................................................................................................TTAGAAGATGACAAGAGCCCTTTGAGAACT........... |
30 |
2 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................................................TCCTGGTTCTGTGGCTCAGAAGCACATTCC..................................................................................... |
30 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................TCTGTAGTCTCTTTTGGGTCTTGCACTTCCC................................................................................................................................................................................. |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................................................................................................................TCTCCTGATGCTAGGTTGGGTAGAAG.................................. |
26 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................................................................................................................................TGCTAGGTTGGTTAGAAGATGACAATAGC....................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................................TGGGCTACTCTGTTTAGTCTAAAACC.................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................................................................................................................................................TGCTAGGTTGGTTAGAAGATGACAATAGCCCT.................... |
32 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................................................................................................................TAGGTTGGTTAGAAGATGACAATAGCCCTTT.................. |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................................................................................................................................................TGCTAGGTTGGTTAGAAGATGACAATAGCCT..................... |
31 |
1 |
1 |
1.00 |
1 |
1 |
| ........................................................................................................................................TTCTGTGGCTCAGAAGCACATTCCAGGGCTT.............................................................................. |
31 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................................................................................................................................................................................TTAGAAGATGACAATAGCCCTTTGAGAACT........... |
30 |
1 |
1 |
1.00 |
1 |
1 |
| ...................................................................................TGCTTGTGGGCTACTCTGTTTAGTCTAAAATT.................................................................................................................................. |
32 |
2 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................................................................................................................TAGGTTGGTTAGAAGATGACAATAGCC...................... |
27 |
0 |
1 |
1.00 |
1 |
1 |