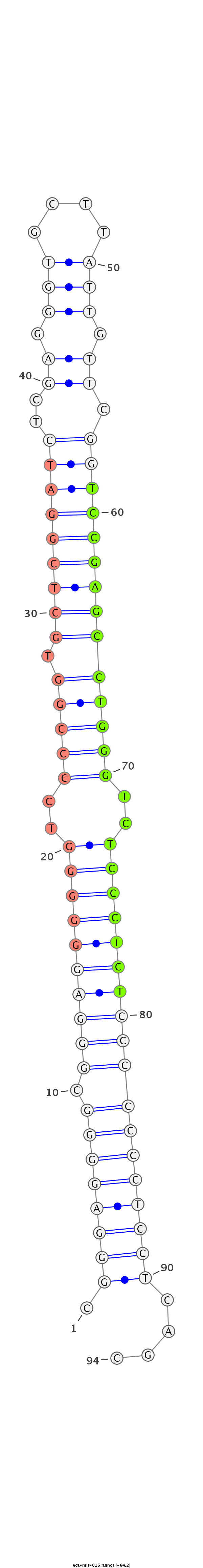
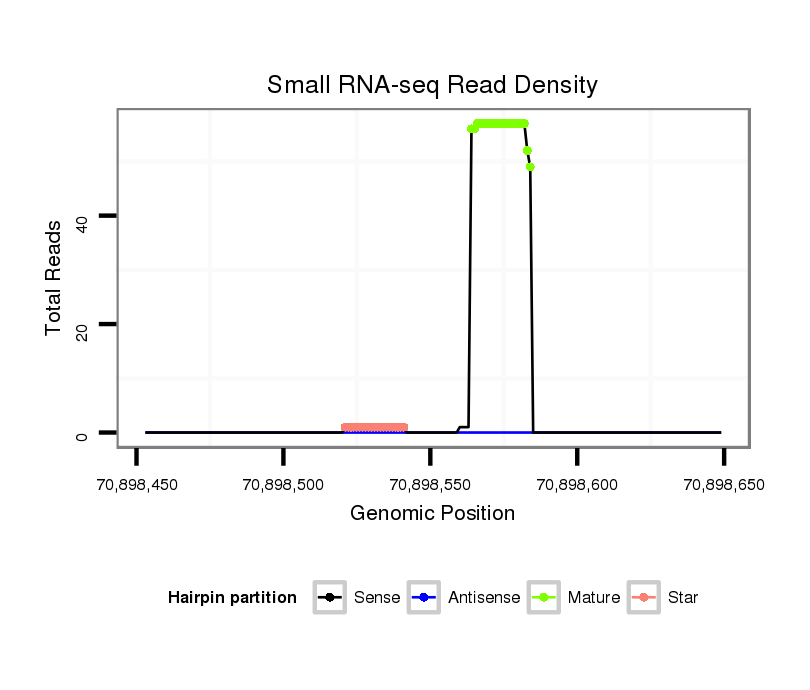
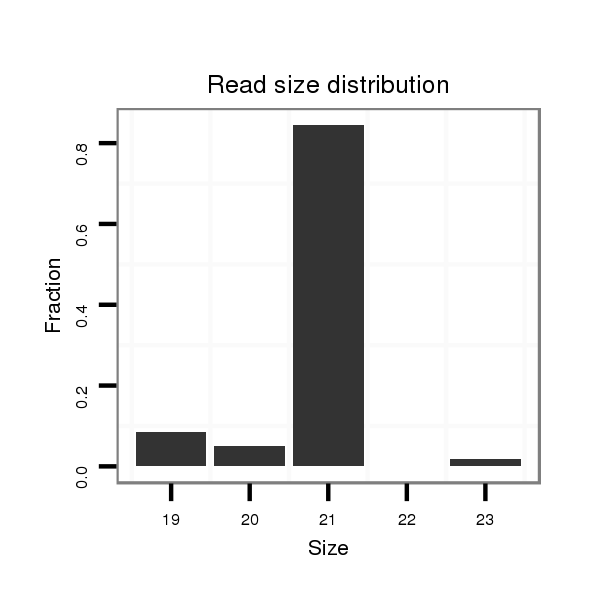
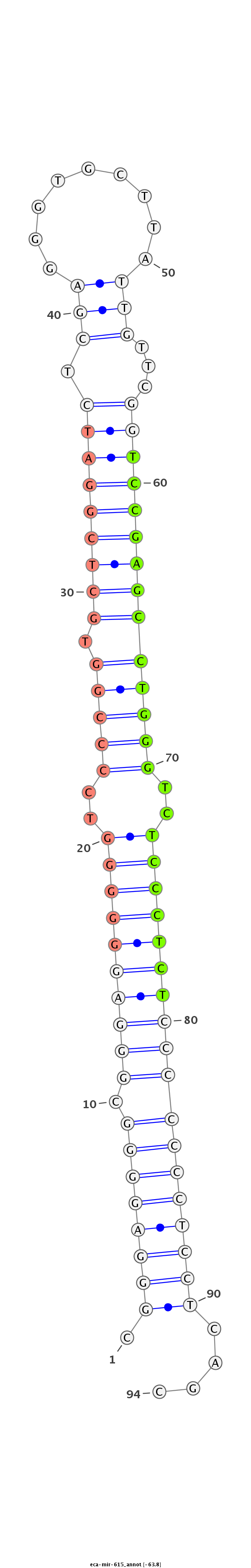
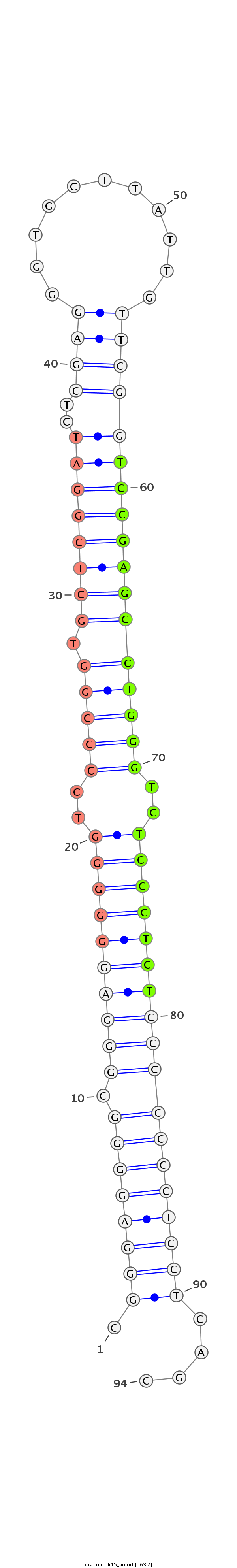
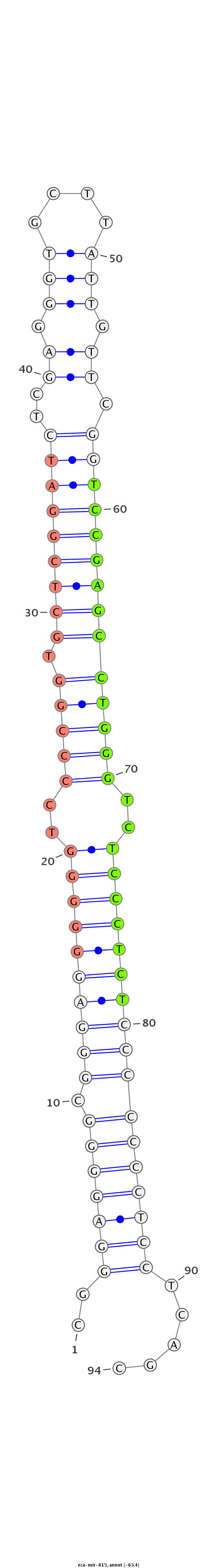
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCT................................................................. |
21 |
0 |
1 |
48.00 |
48 |
48 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTT................................................................ |
22 |
1 |
1 |
18.00 |
18 |
18 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTA................................................................ |
22 |
1 |
1 |
5.00 |
5 |
5 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCT................................................................... |
19 |
0 |
1 |
4.00 |
4 |
4 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTA............................................................... |
23 |
2 |
1 |
3.00 |
3 |
3 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTC.................................................................. |
20 |
0 |
1 |
3.00 |
3 |
3 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTG................................................................ |
22 |
1 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTAT............................................................... |
23 |
2 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCA................................................................. |
21 |
1 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCAA................................................................ |
22 |
2 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTTT.............................................................. |
24 |
3 |
2 |
1.50 |
3 |
3 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTAT................................................................. |
21 |
1 |
1 |
1.00 |
1 |
1 |
| ......................................................................................................ATTGTTCGGTCCGAGCCTGGGTCTCCCTCTTTT.............................................................. |
33 |
3 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTAA.............................................................. |
24 |
3 |
2 |
1.00 |
2 |
2 |
| ...........................................................................................................TCGGTCCGAGCCTGGGTCTCCCT................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTCA.............................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTT............................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTAA............................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................................................................CGAGCCTGGGTCTCCCTCT................................................................. |
19 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................GGGGGTCCCCGGTGCTCGGAT............................................................................................................ |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGGCTCCCTCT................................................................. |
21 |
1 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCCT................................................................ |
22 |
2 |
3 |
0.67 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTAA................................................................. |
21 |
2 |
3 |
0.67 |
2 |
2 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTTAG.............................................................. |
24 |
3 |
2 |
0.50 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTGA............................................................... |
23 |
2 |
2 |
0.50 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTAAA.............................................................. |
24 |
3 |
2 |
0.50 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCACTCTT................................................................ |
22 |
2 |
2 |
0.50 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTCTATT.............................................................. |
24 |
3 |
3 |
0.33 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTAG................................................................. |
21 |
2 |
4 |
0.25 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTAAA................................................................ |
22 |
3 |
9 |
0.11 |
1 |
1 |
| ...............................................................................................................TCCGAGCCTGGGTCTCCCTAAG................................................................ |
22 |
3 |
16 |
0.06 |
1 |
1 |