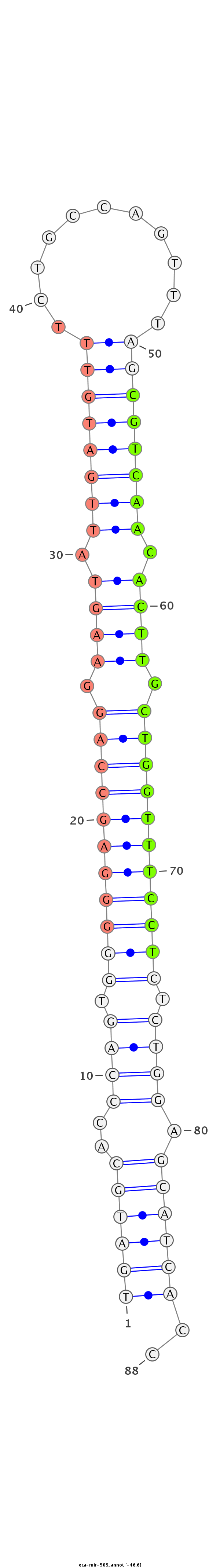
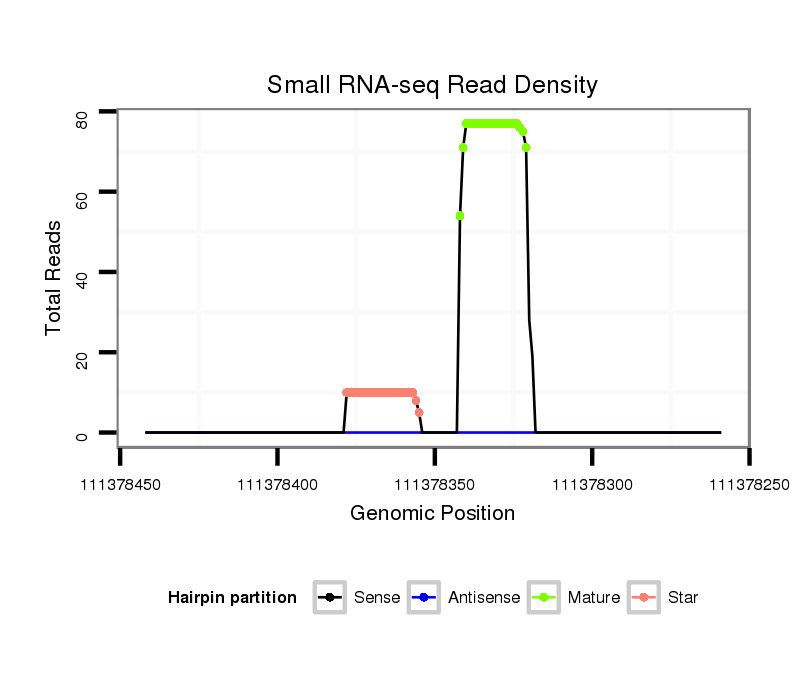
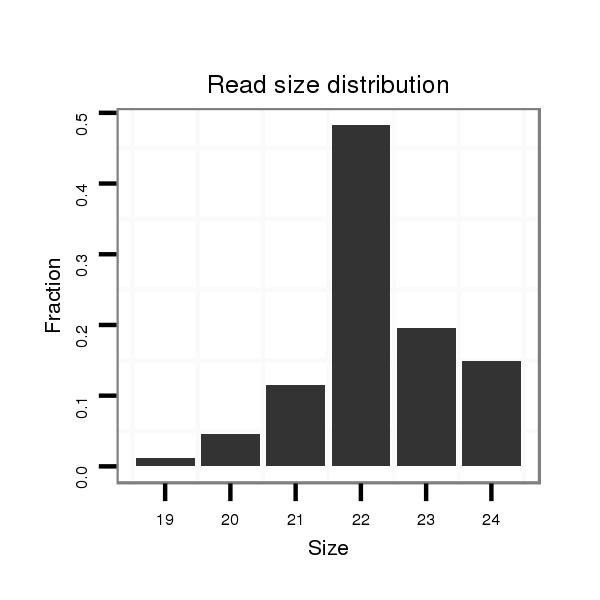
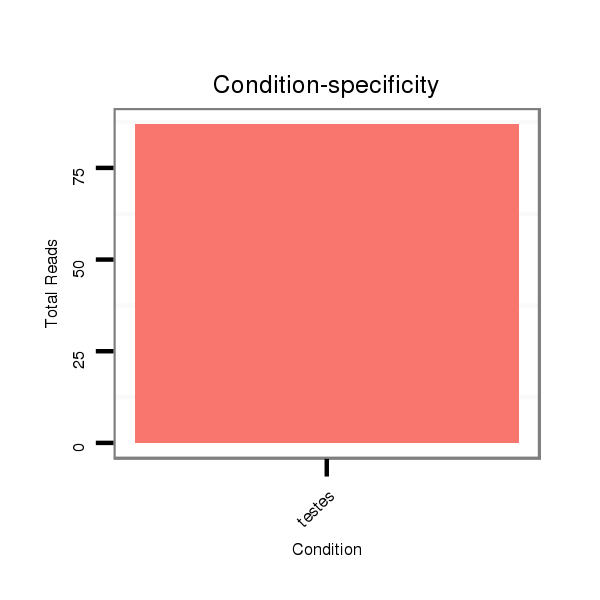
| ....................................................................................................CGTCAACACTTGCTGGTTTCCT.............................................................. |
22 |
0 |
1 |
34.00 |
34 |
34 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCT............................................................ |
24 |
0 |
1 |
8.00 |
8 |
8 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTCT............................................................ |
23 |
0 |
1 |
8.00 |
8 |
8 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCT.............................................................. |
21 |
0 |
1 |
6.00 |
6 |
6 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTC............................................................. |
23 |
0 |
1 |
6.00 |
6 |
6 |
| ................................................................GGGAGCCAGGAAGTATTGATGTTT................................................................................................ |
24 |
0 |
1 |
5.00 |
5 |
5 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCC............................................................... |
21 |
0 |
1 |
4.00 |
4 |
4 |
| ................................................................GGGAGCCAGGAAGTATTGATGTT................................................................................................. |
23 |
0 |
1 |
3.00 |
3 |
3 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTT............................................................. |
23 |
1 |
1 |
3.00 |
3 |
3 |
| ......................................................................................................TCAACACTTGCTGGTTTCCTCT............................................................ |
22 |
0 |
1 |
3.00 |
3 |
3 |
| ......................................................................................................TCAACACTTGCTGGTTTCCT.............................................................. |
20 |
0 |
1 |
3.00 |
3 |
3 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTC............................................................. |
22 |
0 |
1 |
3.00 |
3 |
3 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCTAAA......................................................... |
27 |
3 |
1 |
2.00 |
2 |
2 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTCC............................................................ |
23 |
1 |
1 |
2.00 |
2 |
2 |
| ................................................................GGGAGCCAGGAAGTATTGATGT.................................................................................................. |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................................................................TCAACACTTGCTGGTTTCCTCC............................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCTTAT......................................................... |
27 |
3 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCAT........................................................... |
25 |
2 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTCTAA.......................................................... |
25 |
2 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCATT.......................................................... |
26 |
2 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTT................................................................. |
19 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................GTCAACACTTGCTGGTTTCGTCT............................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................CGTGGCGTCAACACTTGCTGGTTTCCT.............................................................. |
27 |
3 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGGTGGTTTCCT.............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTC................................................................ |
20 |
0 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTG............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTCCTCTAAT......................................................... |
27 |
3 |
1 |
1.00 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTTACT.............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| .....................................................................................................GTCAACGCTCGCTGGTTTCATCT............................................................ |
23 |
3 |
2 |
0.50 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTCCCTCAT........................................................... |
25 |
3 |
2 |
0.50 |
1 |
1 |
| .....................................................................................................GTCAACACTTGCTGGTTTCCTTT............................................................ |
23 |
1 |
3 |
0.33 |
1 |
1 |
| ....................................................................................................CGTCAACACTTGCTGGTTAA................................................................ |
20 |
2 |
7 |
0.14 |
1 |
1 |