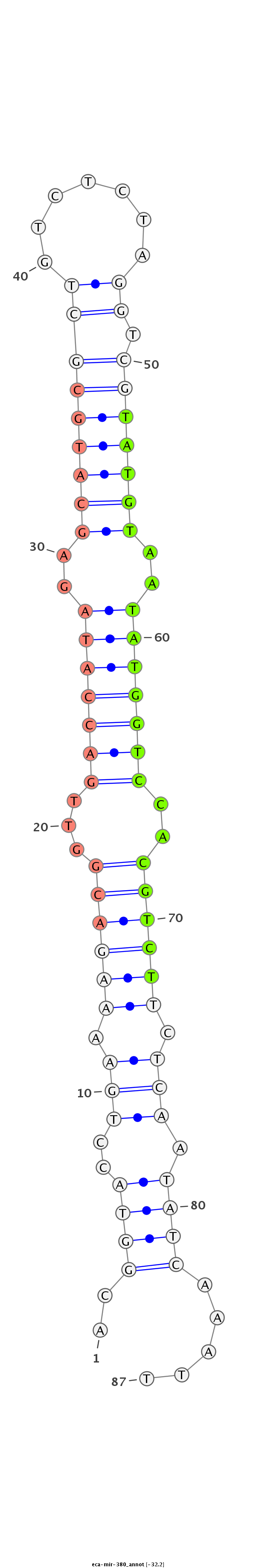
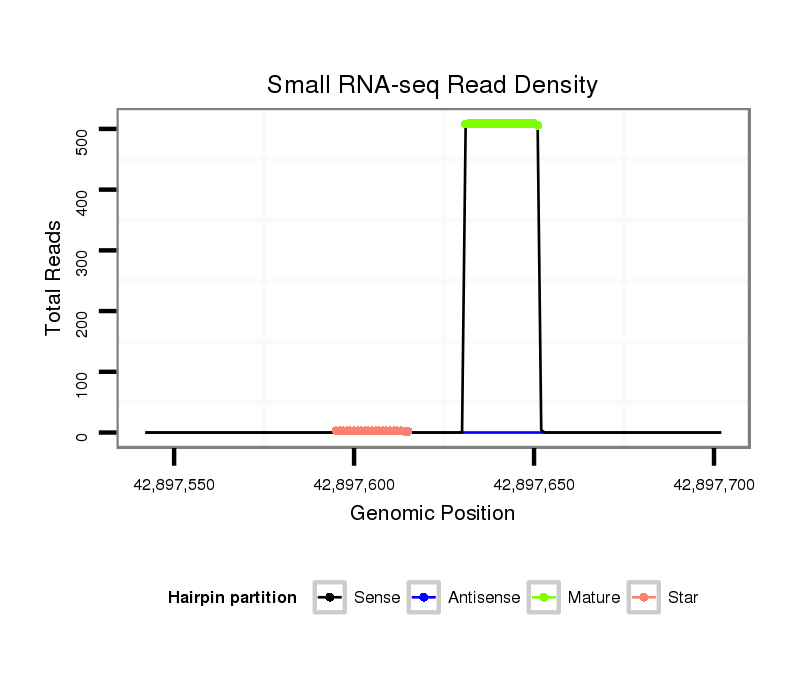
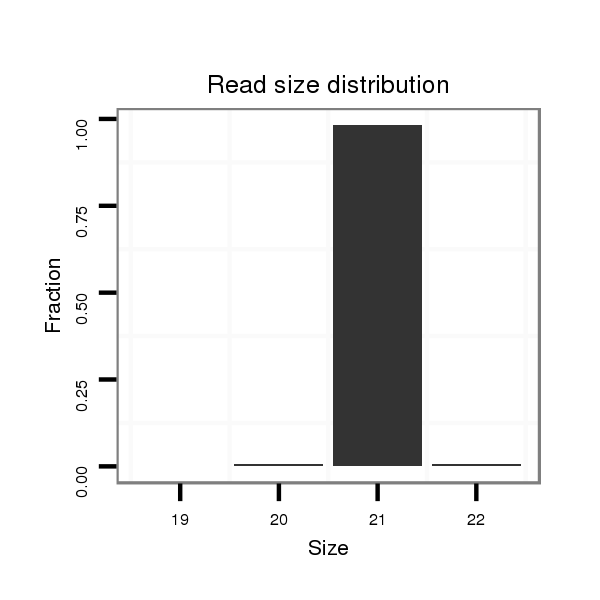
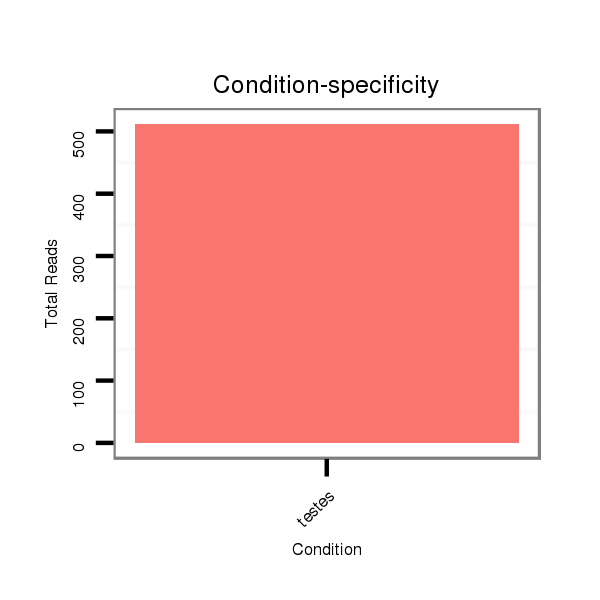
ID:eca-mir-380 |
Coordinate:chr24:42897592-42897652 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
TGAGGAAAGAGACGCCGGTTCTGACATCAGCCCTTTCCACGGTACCTGAAAAGACGGTTGACCATAGAGCATGCGCTGTCTCTAGGTCGTATGTAATATGGTCCACGTCTTCTCAATATCAAATTCAGTCGCAGAGGACATCCCAGAGGCAAAACCAACTT
**************************************..((((..(((.((((((...(((((((..(((((((((.......)).)))))))..)))))))..)))))).))).)))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .........................................................................................TATGTAATATGGTCCACGTCT................................................... | 21 | 0 | 1 | 501.00 | 501 | 501 |
| .........................................................................................TATGTAATATGGGCCACGTCT................................................... | 21 | 1 | 1 | 4.00 | 4 | 4 |
| .........................................................................................TATGTAATATGGTCCACGTAT................................................... | 21 | 1 | 1 | 4.00 | 4 | 4 |
| .........................................................................................TATGTAATATGGTCCACGTCTT.................................................. | 22 | 0 | 1 | 4.00 | 4 | 4 |
| .........................................................................................TATGTAATATGGTCCACGTGT................................................... | 21 | 1 | 1 | 4.00 | 4 | 4 |
| .........................................................................................TATGTAATATGGTCCACGTC.................................................... | 20 | 0 | 1 | 3.00 | 3 | 3 |
| .........................................................................................TATGTAATATGGTCCACGTCTGA................................................. | 23 | 2 | 1 | 2.00 | 2 | 2 |
| .....................................................ACGGTTGACCATAGAGCATGC....................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 |
| .........................................................................................TATGTAATATGGTCCACGTCTTGGAA.............................................. | 26 | 3 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGGACCACGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGGTGCACGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .....................................................ACGGTTGACCATAGAGCAT......................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGGTCCACGTCTAG................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGATCCACGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGGTCCAAGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGGTCCACGTCTG.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TGTGTAATATGGTCCACGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................ATGTAATATGGTCCACGTCT................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................ACGGTTGACCATAGAGCATGCGT..................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATATGTTCCACGTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .....................................................CCGGTTGACCATAGAGCATGCGT..................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATAGGGTCCACGTAT................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 |
| .........................................................................................TATGTAATAGGGTCCACGTCTAT................................................. | 23 | 3 | 2 | 0.50 | 1 | 1 |
| .........................................................................................TATGTAATAAGGTCCACGTAT................................................... | 21 | 2 | 3 | 0.33 | 1 | 1 |
| .........................................................................................TATGTACTAGGGTCCACGTCT................................................... | 21 | 2 | 3 | 0.33 | 1 | 1 |
|
ACTCCTTTCTCTGCGGCCAAGACTGTAGTCGGGAAAGGTGCCATGGACTTTTCTGCCAACTGGTATCTCGTACGCGACAGAGATCCAGCATACATTATACCAGGTGCAGAAGAGTTATAGTTTAAGTCAGCGTCTCCTGTAGGGTCTCCGTTTTGGTTGAA
************************************..((((..(((.((((((...(((((((..(((((((((.......)).)))))))..)))))))..)))))).))).)))).....************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
Generated: 09/01/2015 at 11:54 AM