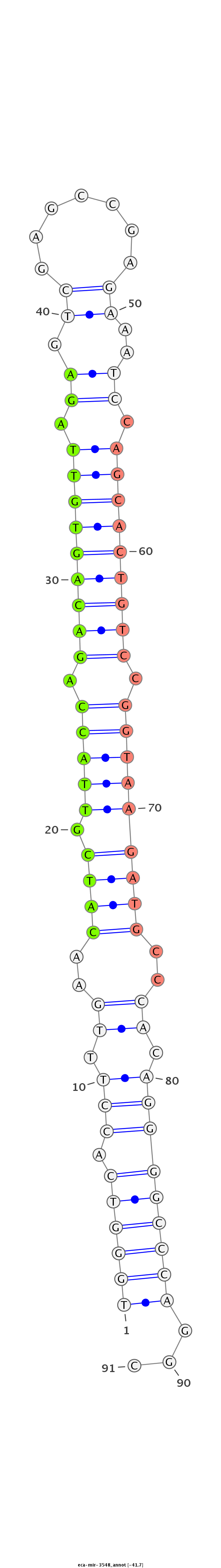
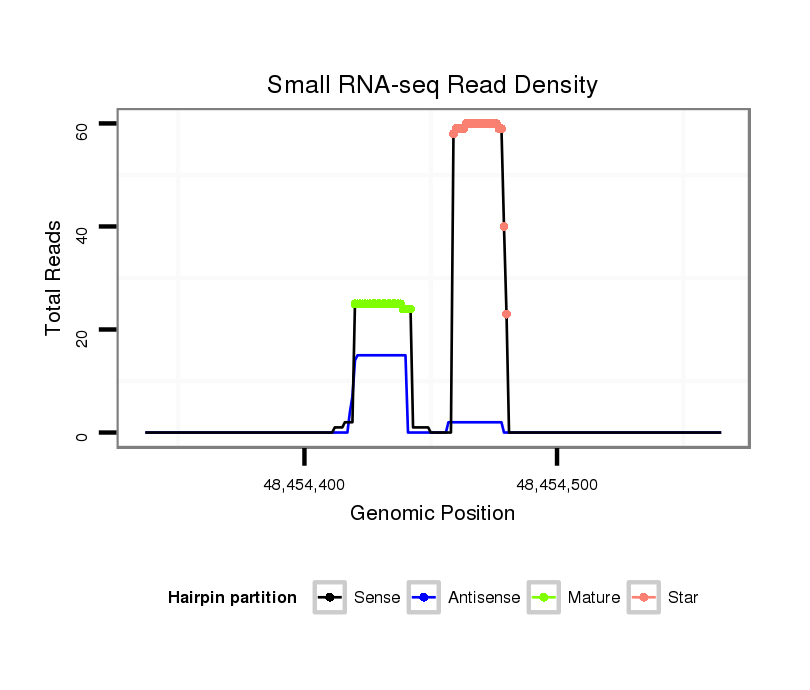
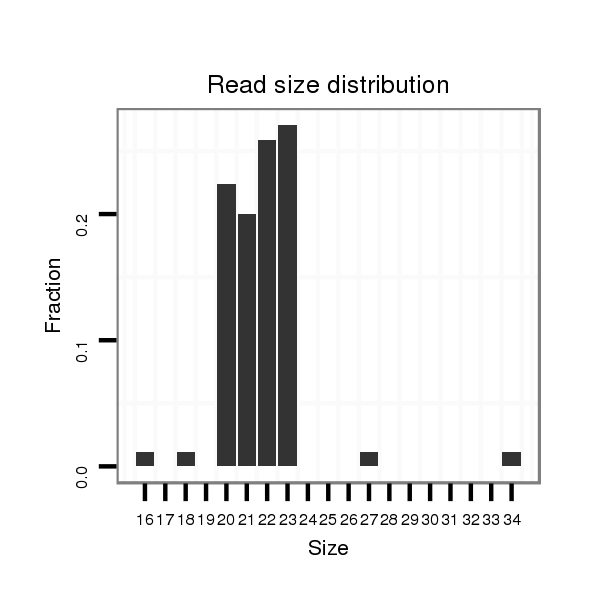
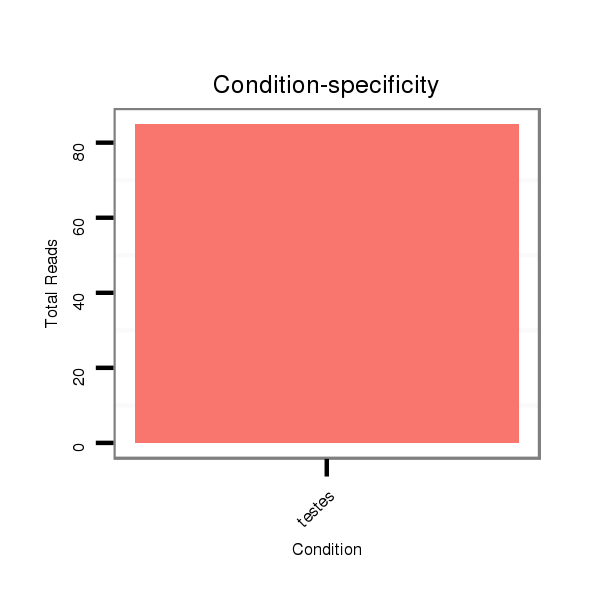
ID:eca-mir-3548 |
Coordinate:chr2:48454387-48454515 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
|
CTTGGTCCATCTCTGGAGTCCGGGAGCTCAGGACCGCGCATCCTCGGTCGCTCCCCAGCGGGCGGCAGTGGGTCACCTTTGAACATCGTTACCAGACAGTGTTAGAGTCGAGCCGAGAAATCCAGCACTGTCCGGTAAGATGCCCACAGGGGCCCAGGCGCCTCAGCCTGAGGACGTGGACGGCATGTGGCACCCGCCCCAGGTAGGCAGGCCCATGTGGAAACAGGAC
********************************************************************((((((.(((.((..((((.(((((.(((((((((.((.((.......))..)).))))))))).)))))))))..)).)))))))))...********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ..........................................................................................................................CAGCACTGTCCGGTAAGATGCCT.................................................................................... | 23 | 1 | 1 | 26.00 | 26 | 26 |
| ...................................................................................CATCGTTACCAGACAGTGTTAGA........................................................................................................................... | 23 | 0 | 1 | 23.00 | 23 | 23 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGATGCC..................................................................................... | 22 | 0 | 1 | 22.00 | 22 | 22 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGATG....................................................................................... | 20 | 0 | 1 | 19.00 | 19 | 19 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGATGC...................................................................................... | 21 | 0 | 1 | 16.00 | 16 | 16 |
| ...................................................................................CATCGTTACCGGACAGTGTTAGA........................................................................................................................... | 23 | 1 | 1 | 4.00 | 4 | 4 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGATGCCA.................................................................................... | 23 | 1 | 1 | 3.00 | 3 | 3 |
| ..........................................................................................................................TAGCACTGTCCGGTAAGATGCC..................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 |
| ...................................................................................CATCGTTACCAGACAGTGTTAGT........................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 |
| ...............................................................................................................................CTGTCCGGTAAGATGC...................................................................................... | 16 | 0 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGATGT...................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| ...................................................................................CATCGTTACCGGACAGTGTTGGA........................................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 |
| ..........................................................................................................................CAGCACTGTCCGGTAAGA......................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................................................................TGAACATCGTTACCAGACAGTGTTAGAGTCGAGC.................................................................................................................... | 34 | 0 | 1 | 1.00 | 1 | 1 |
| ...................................................................................CATCGTTACCAGACAGTGTTAGAA.......................................................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 |
| ...........................................................................CCTTTGAACATCGTTACCAGACAGTGT............................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ...........................................................................................................................AGCACTGTCCGGTAAGATGCC..................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 |
|
GAACCAGGTAGAGACCTCAGGCCCTCGAGTCCTGGCGCGTAGGAGCCAGCGAGGGGTCGCCCGCCGTCACCCAGTGGAAACTTGTAGCAATGGTCTGTCACAATCTCAGCTCGGCTCTTTAGGTCGTGACAGGCCATTCTACGGGTGTCCCCGGGTCCGCGGAGTCGGACTCCTGCACCTGCCGTACACCGTGGGCGGGGTCCATCCGTCCGGGTACACCTTTGTCCTG
**********************************************************************((((((.(((.((..((((.(((((.(((((((((.((.((.......))..)).))))))))).)))))))))..)).)))))))))...******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...................................................................................GTAGCAATGGTCTGTCACAAT............................................................................................................................. | 21 | 0 | 1 | 7.00 | 7 | 7 |
| .................................................................................TTGTAGCAATGGTCTGTCACAAT............................................................................................................................. | 23 | 0 | 1 | 4.00 | 4 | 4 |
| ..................................................................................TGTAGCAATGGTCTGTCACAAT............................................................................................................................. | 22 | 0 | 1 | 3.00 | 3 | 3 |
| ........................................................................................................................AGGTCGTGACAGGCCATTCTAC....................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 |
| ................................................................................TTTGTAGCAATGGTCTGTCACAAT............................................................................................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 |
| ....................................................................................TAGCAATGGTCTGTCACAAT............................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 |
| ...................................................................................GTAGGAGTGGTCTGTCACAAT............................................................................................................................. | 21 | 2 | 9 | 0.11 | 1 | 1 |
Generated: 09/01/2015 at 11:48 AM