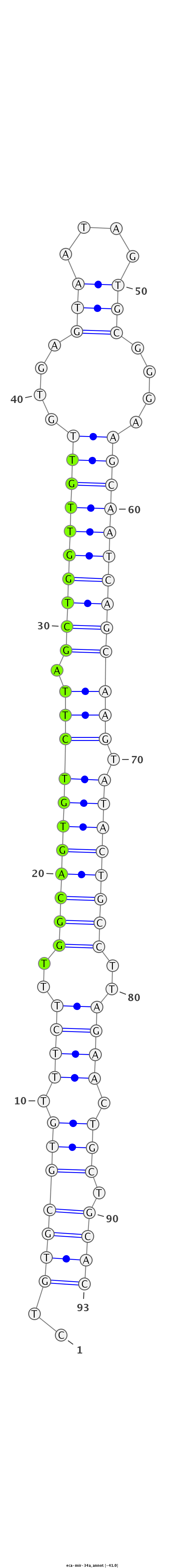
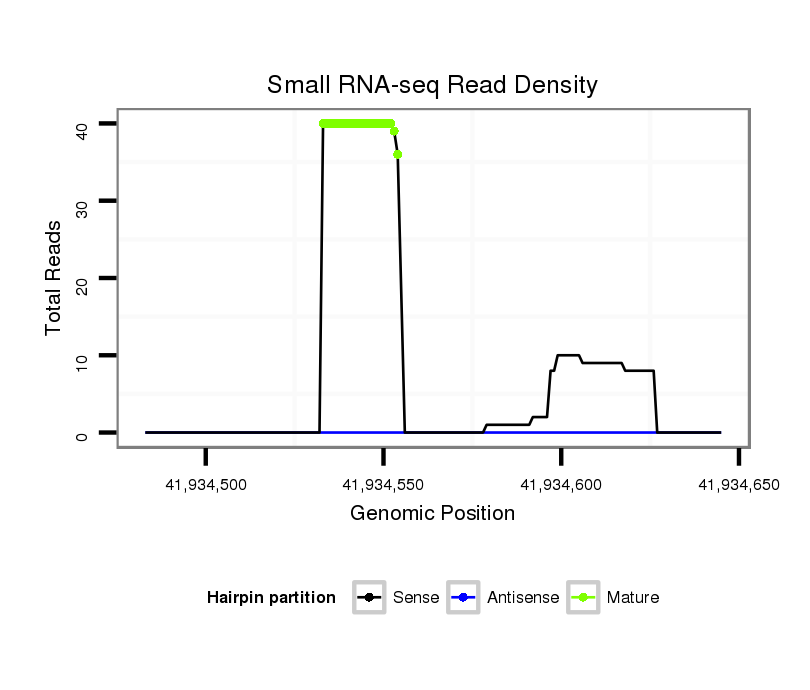
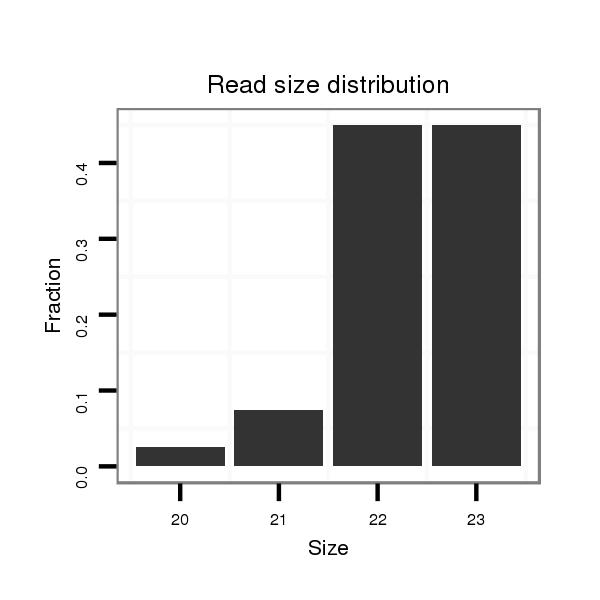
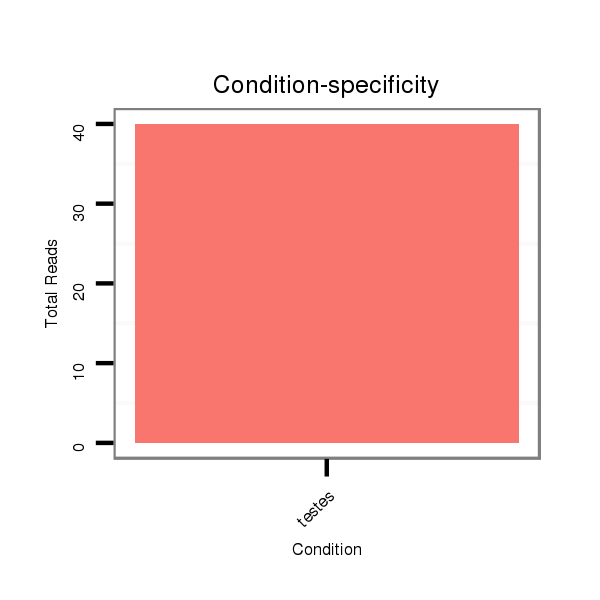
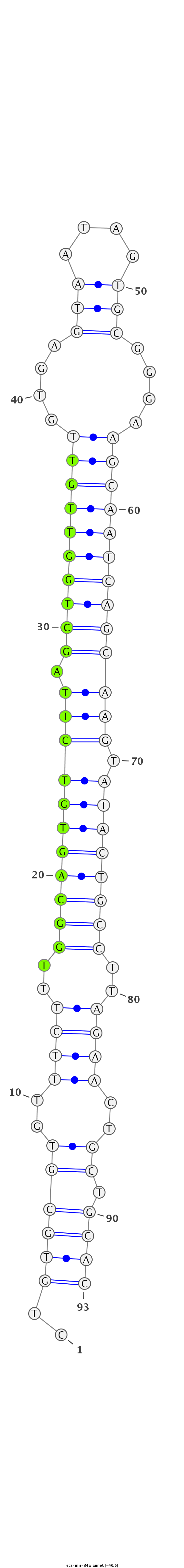
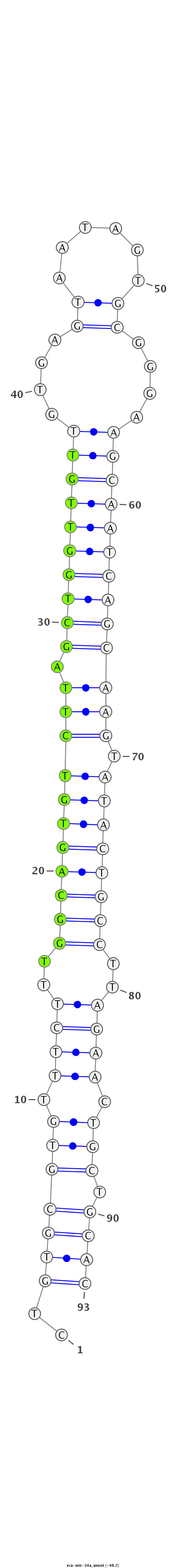
ID:eca-mir-34a |
Coordinate:chr2:41934533-41934595 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.6 | -40.5 | -40.2 |
 |
 |
 |
|
GCCGGGTGTGCCTGCTGTGCCACGGACAGGGCCAGCTGTGCGTGTTTCTTTGGCAGTGTCTTAGCTGGTTGTTGTGAGTAATAGTGCGGGAAGCAATCAGCAAGTATACTGCCTTAGAACTGCTGCACGTTGTGGGGCCCGAGAGGAAGGGACAGGTGAGAGA
***********************************..(((((((.((((..(((((((((((.((((((((((....(((....)))....))))))))))))).))))))))..)))).))).))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ..................................................TGGCAGTGTCTTAGCTGGTTGT........................................................................................... | 22 | 0 | 1 | 18.00 | 18 | 18 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTT.......................................................................................... | 23 | 0 | 1 | 18.00 | 18 | 18 |
| ..................................................................................................................TAGAACTGCTGCACGTTGTGGGGCCCGAGA................... | 30 | 0 | 1 | 6.00 | 6 | 6 |
| ..................................................TGGCAGTGTCTTAGCTGGTTG............................................................................................ | 21 | 0 | 1 | 3.00 | 3 | 3 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTTT......................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 |
| ....................................................................................................................GAACTGCTGCACGTTGTGGGGCCCGAGA................... | 28 | 0 | 1 | 2.00 | 2 | 2 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTA.......................................................................................... | 23 | 1 | 1 | 2.00 | 2 | 2 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTAA......................................................................................... | 24 | 2 | 1 | 2.00 | 2 | 2 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTAGT........................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................TGCCTTAGAACTGCTGCACGTTGTGGGGCT........................ | 30 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTTTT........................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTT............................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTAT......................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGTTAT........................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTTT............................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 |
| ................................................................................................TCAGCAAGTATACTGCCTTAGAACTGC........................................ | 27 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................................................................................TAGAACTGCTGCACGTTGTGGGGCCCGAGTA.................. | 31 | 2 | 1 | 1.00 | 1 | 1 |
| .............................................................................................................TGCCTTAGAACTGCTGCACGTTGTGG............................ | 26 | 0 | 1 | 1.00 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGGTTGAAA......................................................................................... | 24 | 3 | 3 | 0.33 | 1 | 1 |
| ..................................................TGGCAGTGTCTTAGCTGA............................................................................................... | 18 | 1 | 13 | 0.08 | 1 | 1 |
|
CGGCCCACACGGACGACACGGTGCCTGTCCCGGTCGACACGCACAAAGAAACCGTCACAGAATCGACCAACAACACTCATTATCACGCCCTTCGTTAGTCGTTCATATGACGGAATCTTGACGACGTGCAACACCCCGGGCTCTCCTTCCCTGTCCACTCTCT
***********************************..(((((((.((((..(((((((((((.((((((((((....(((....)))....))))))))))))).))))))))..)))).))).))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ...........................................................................................TCGTTAGTCGATGATGTGACGGA................................................. | 23 | 3 | 4 | 0.25 | 1 | 1 |
Generated: 09/01/2015 at 11:48 AM