
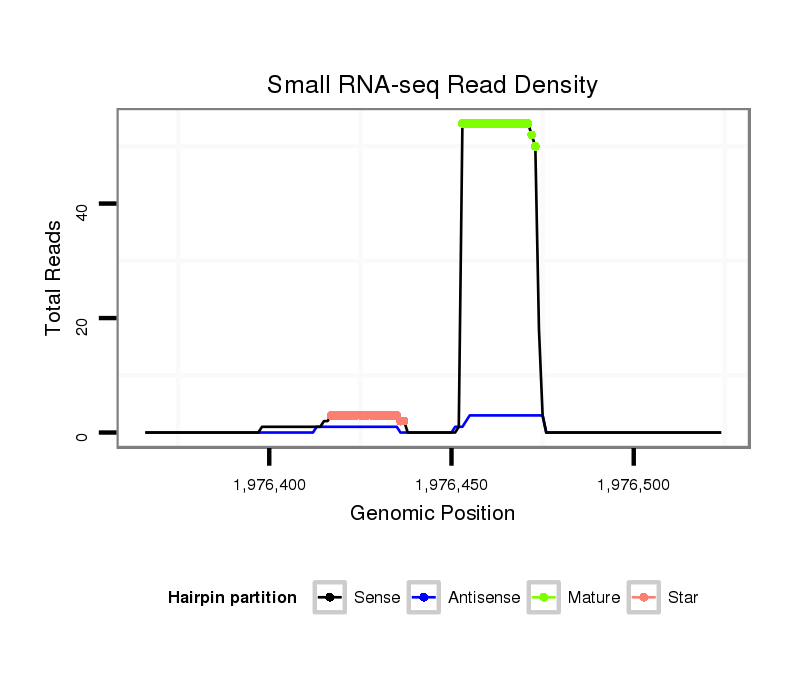
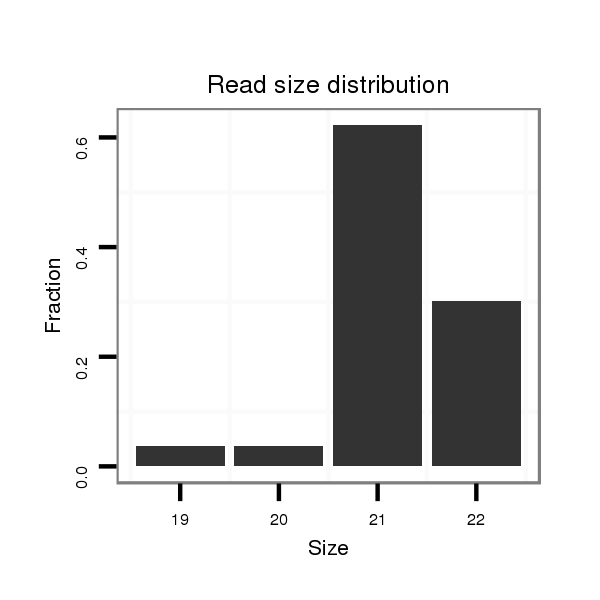
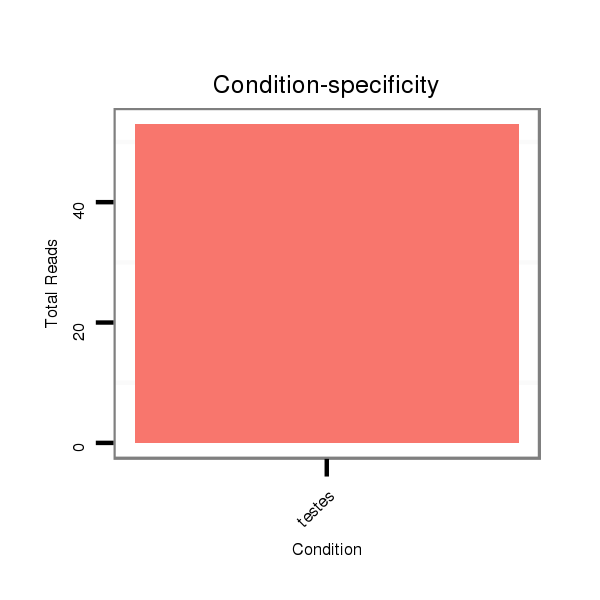
ID:eca-mir-338 |
Coordinate:chr11:1976416-1976474 + |
Confidence:Known |
Class:Unknown |
Genomic Locale:intergenic |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.2 | -40.1 | -40.1 |
 |
 |
 |
|
GTGATGGACGTGGCCCCGCCAGCCCCAGCCGCCGCACGGGCCGTCCTCCCCAACAATATCCTGGTGCTGAGTGATGACACACGCAACTCCAGCATCAGTGATTTTGTTGAAGAGGGCAGCTGCCAGCCTCCTGACCTGCCTGCCGCGCCCACCCAGCCC
************************************..(((.((((((..((((((.(((((((((((((((..((.....))..))).))))))))).))).))))))..)))))).)))..************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| .......................................................................................TCCAGCATCAGTGATTTTGTT................................................... | 21 | 0 | 1 | 31.00 | 31 | 31 |
| .......................................................................................TCCAGCATCAGTGATTTTGTTG.................................................. | 22 | 0 | 1 | 15.00 | 15 | 15 |
| .......................................................................................TCCAGCATCAGTGATTTTGTTGA................................................. | 23 | 0 | 1 | 3.00 | 3 | 3 |
| .......................................................................................TCCAGCATCAGTGATTTTG..................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................TCCAGCATCAGTGATTTTGT.................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................TCCAGCATCAGTGATTTTGTA................................................... | 21 | 1 | 1 | 2.00 | 2 | 2 |
| ...................................................AACAATATCCTGGTGCTGAGT....................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 |
| .......................................................................................TCCAGCATCAGTAATTTTGTT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 |
| .......................................................................................TCCAGCGTCAGTGATTTTGTTG.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 |
| .................................................CCAACAATATCCTGGTGCTGA......................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................................TCCAGCATCAGTGATTTTGTTAA................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 |
| ......................................................................................CTCCAGCATCAGTGATTTTGTT................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 |
| ................................................CCCAACAATATCCTGGTGCTGT......................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 |
| ................................CGCACGGGCCGTCCTCCCC............................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................................TCCAGCATCAGTGATTTTGTTT.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 |
| .......................................................................................TCCAGCATCAGTGATTTTGTAA.................................................. | 22 | 2 | 6 | 0.17 | 1 | 1 |
|
CACTACCTGCACCGGGGCGGTCGGGGTCGGCGGCGTGCCCGGCAGGAGGGGTTGTTATAGGACCACGACTCACTACTGTGTGCGTTGAGGTCGTAGTCACTAAAACAACTTCTCCCGTCGACGGTCGGAGGACTGGACGGACGGCGCGGGTGGGTCGGG
************************************..(((.((((((..((((((.(((((((((((((((..((.....))..))).))))))))).))).))))))..)))))).)))..************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR871532 testes |
|---|---|---|---|---|---|---|
| ..............................................TGGGGTTGTTATAGGACCACGACT......................................................................................... | 24 | 1 | 1 | 2.00 | 2 | 2 |
| ...............................................GGGGTTGTTATAGGACCACGACT......................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 |
| ......................................................................................AAGGTCGTAGTCACTAAAACAACT................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 |
| .....................................................................................TGAGGTCGTAGTCACTAAAACAACT................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 |
| .......................................................................................AGGTCGGAGTCACTAAAACAACT................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 |
| ........................................................................................GGTCGTAGTCACTAAAACAACT................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 |
| .........................................................................................GTCGTAGTCACTAAAACAACT................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 |
| .....................................................................................TGATGTCGTAGTCACTAAAACAACT................................................. | 25 | 1 | 1 | 1.00 | 1 | 1 |
Generated: 09/01/2015 at 11:34 AM