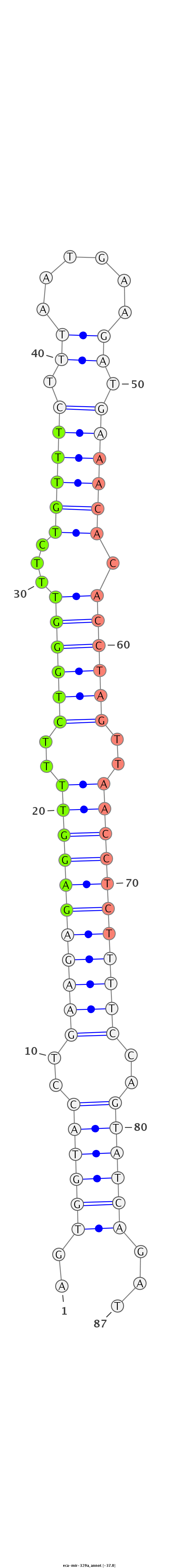
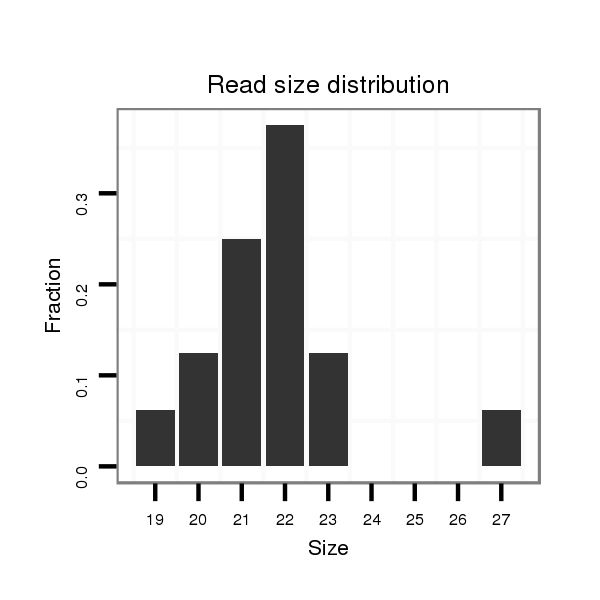
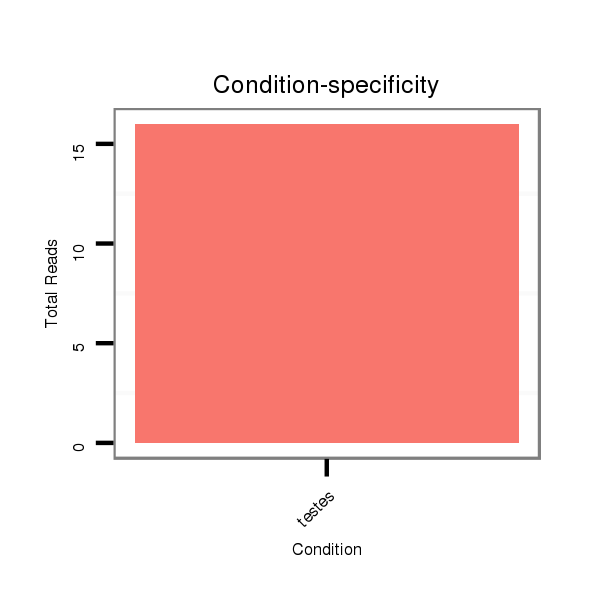
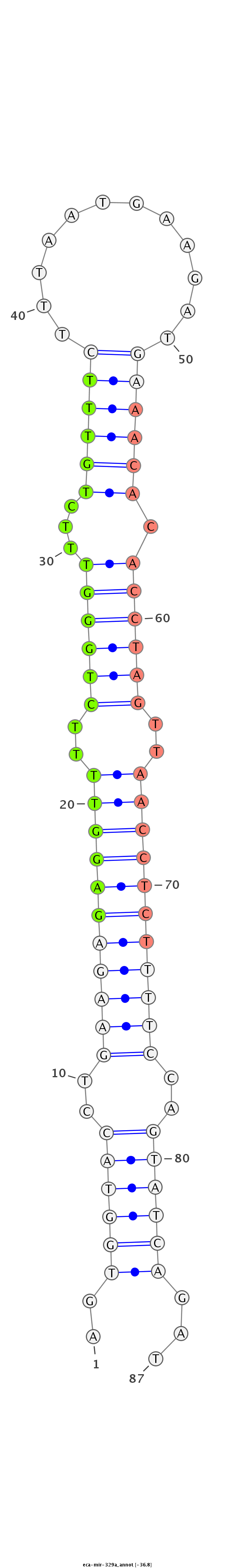
| ...............................................................................................................AACACACCTAGTTAACCTCTCT................................................................. |
22 |
1 |
1 |
14.00 |
14 |
14 |
| ...............................................................................................................AACACACCTAGTTAACCTCTCC................................................................. |
22 |
2 |
1 |
11.00 |
11 |
11 |
| ................................................................................................................ACACACCTAGTTAACCTCTCCT................................................................ |
22 |
2 |
1 |
5.00 |
5 |
5 |
| ..........................................................................GAGGTTTTCTGGGTTTCTGTTT...................................................................................................... |
22 |
0 |
1 |
3.00 |
3 |
3 |
| .........................................................................AGAGGTTTTCTGGGTTTCTGTT....................................................................................................... |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ..........................................................................GAGGTTTTCTGGGTTTCTGTTTC..................................................................................................... |
23 |
0 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................AACACACCTAGTTAACCTCTC.................................................................. |
21 |
1 |
1 |
2.00 |
2 |
2 |
| ................................................................................................................ACACACCTAGTTAACCTCTATT................................................................ |
22 |
1 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................AACACACCTAGTTAACCTCT................................................................... |
20 |
0 |
1 |
2.00 |
2 |
2 |
| ...............................................................................TTTCTGGGTTTCTGTTTCTTT.................................................................................................. |
21 |
0 |
1 |
2.00 |
2 |
2 |
| ...............................................................................................................AACACACCTAGTTAACCTC.................................................................... |
19 |
0 |
1 |
1.00 |
1 |
1 |
| ................................................................................................................ACACACCTAGTTAACCTCTTT................................................................. |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................AACACACCTAGTTAACCTCTCTA................................................................ |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................AACACACCTAGTTAACCTCTCTTT............................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................AACACACCTAGTTAACCTCTTAA................................................................ |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................AACACACCTAGTTAACCTCTT.................................................................. |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................AACACACCTAGTTAACCTCTTT................................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
| .........................................................................AGAGGTTTTCTGGGTTTCTGTTTCTTT.................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
1 |
| ...............................................................................................................CACACACCTAGTTAACCTCTCT................................................................. |
22 |
2 |
4 |
0.25 |
1 |
1 |
| ................................................................................................................ACACACCTAGTTAACCTCTCCA................................................................ |
22 |
3 |
8 |
0.13 |
1 |
1 |