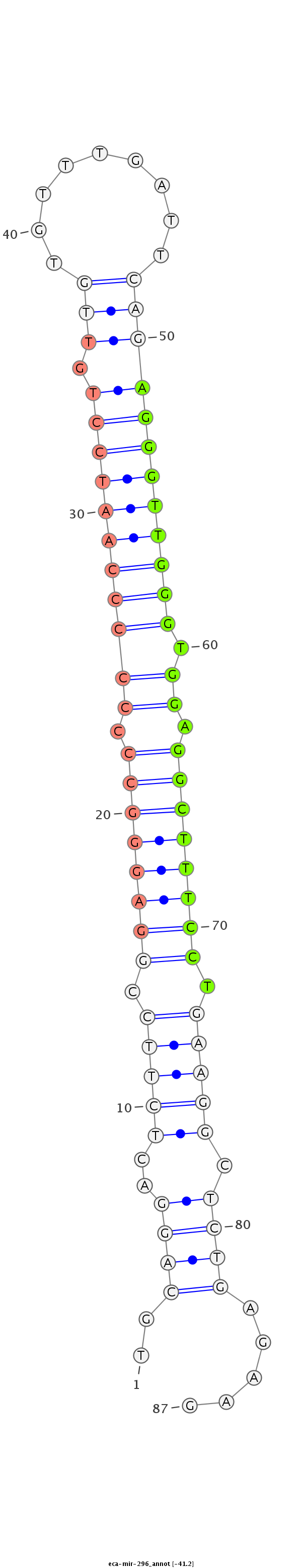
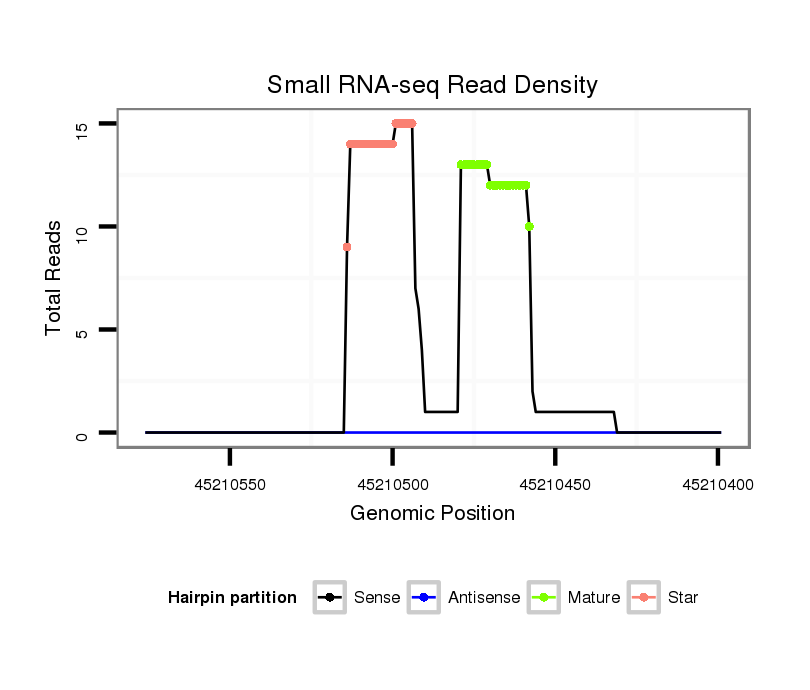
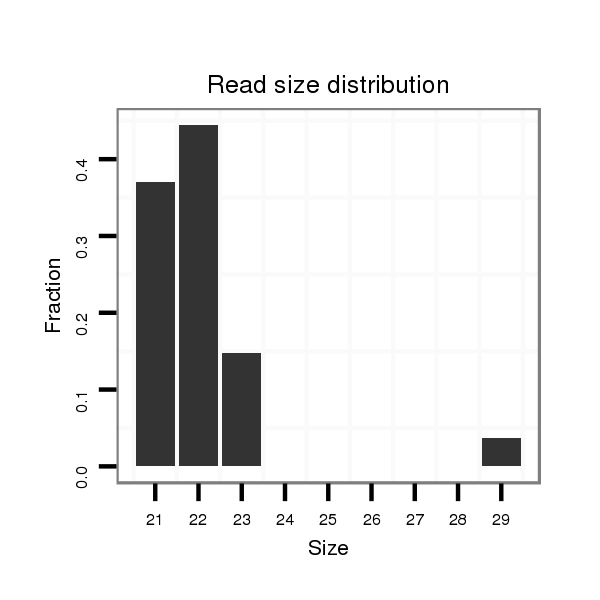
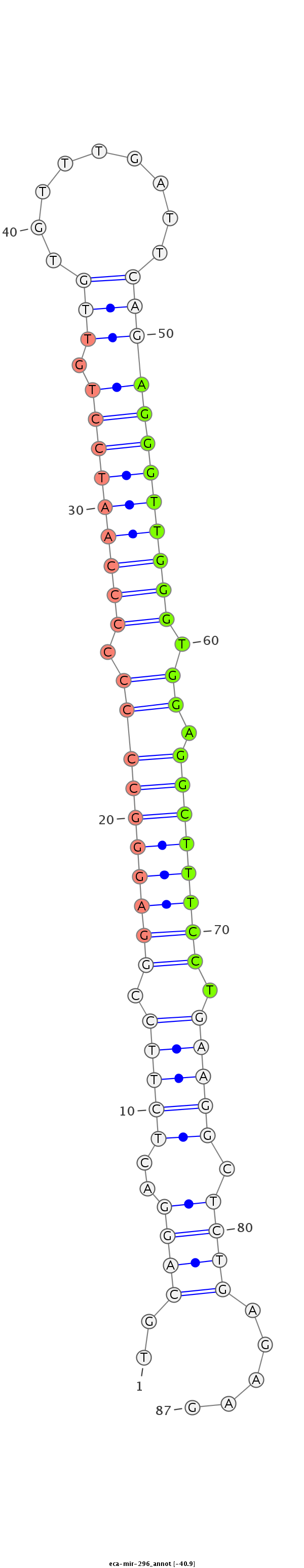
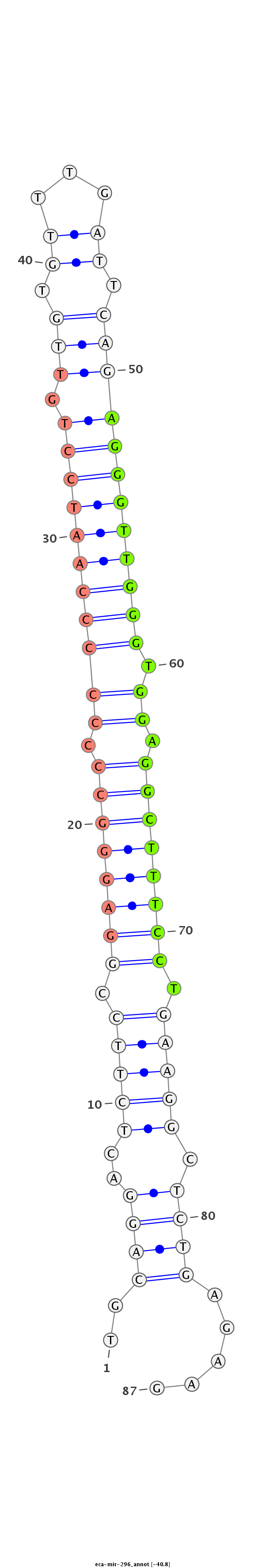
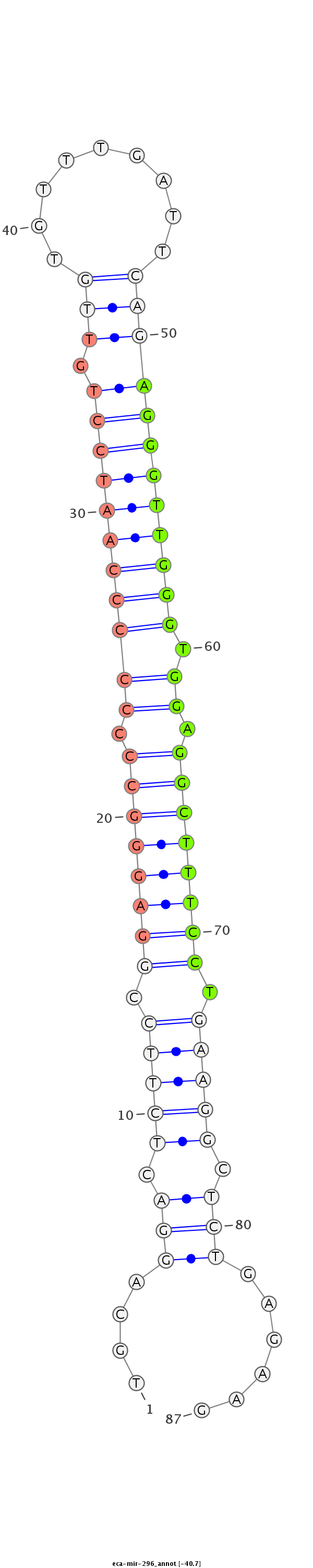
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCT........................................................... |
22 |
0 |
1 |
9.00 |
9 |
9 |
| ..............................................................GAGGGCCCCCCCCAATCCTGT............................................................................................... |
21 |
0 |
1 |
8.00 |
8 |
8 |
| ...............................................................AGGGCCCCCCCCAATCCTGTTGT............................................................................................ |
23 |
0 |
1 |
3.00 |
3 |
3 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCA........................................................... |
22 |
1 |
1 |
3.00 |
3 |
3 |
| ..............................................................GAGGGCCCCCCCCAATCCTGC............................................................................................... |
21 |
1 |
2 |
2.00 |
4 |
4 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCC............................................................ |
21 |
0 |
1 |
2.00 |
2 |
2 |
| ...............................................................AGGGCCCCCCCCAATCCTGTTG............................................................................................. |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ..............................................................GAGGGCCCCCCCCAATCCTGTT.............................................................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
| ..............................................................GAGGGCCCCCCCCAATCCTGTA.............................................................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ..............................................................GAGGGCCCCCCCCAATCCTGA............................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................................CGGGTTGGGTGGAGGCTTTCCT........................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCAAA......................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| .............................................................................TCCTGTTGTGTTTGATTCAGAGGGTTGGG........................................................................ |
29 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCTAAT........................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGGTGGGTGGAGGCTTTCCTTA......................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCTT.......................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCTG.......................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................................................................................GAAGGCTCTGAGAAGAGCTGCACCCA................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCCTAT......................................................... |
24 |
2 |
1 |
1.00 |
1 |
1 |
| .................................................................................................AGGGTTGGGTGGAGGCTTTCTTAAA........................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |