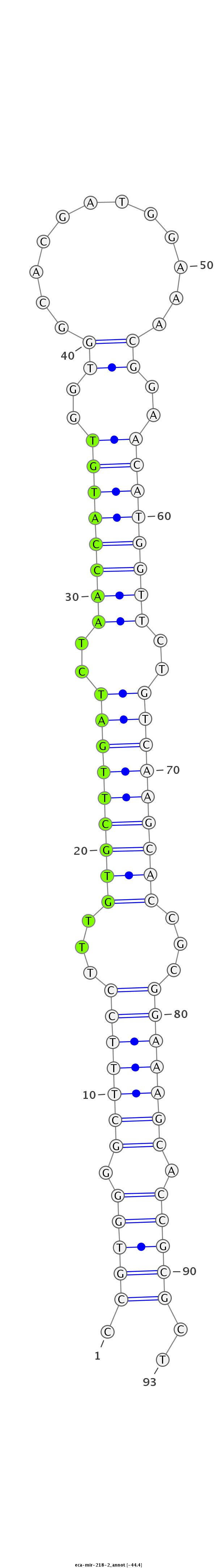
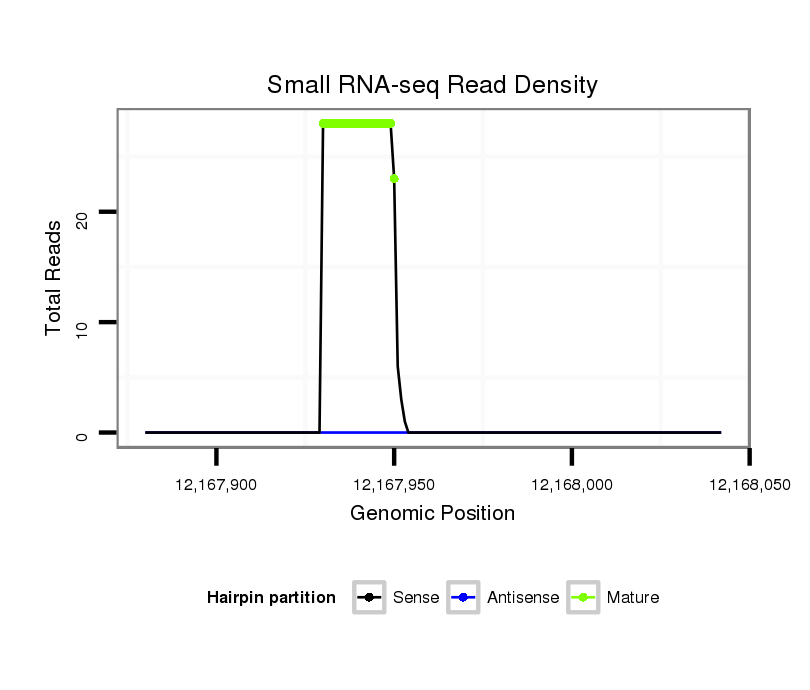
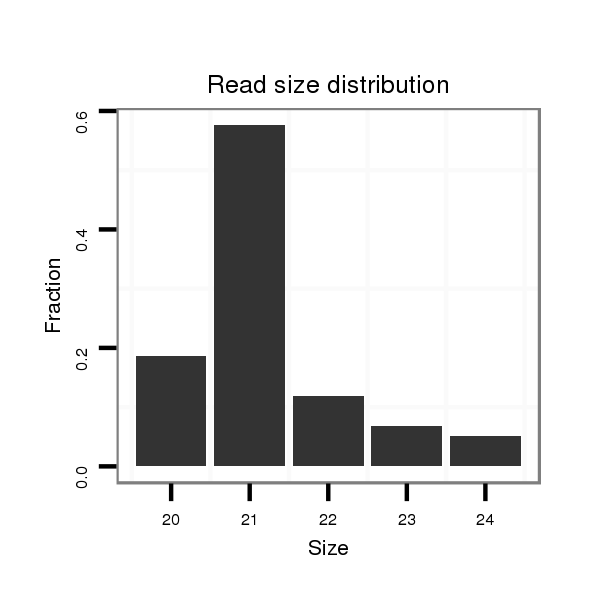
| ..................................................TTGTGCTTGATCTAACCATGT............................................................................................ |
21 |
0 |
2 |
17.00 |
34 |
34 |
| ..................................................TTGTGCTTGATCTAACCATG............................................................................................. |
20 |
0 |
2 |
5.50 |
11 |
11 |
| ..................................................TTGTGCTTGATCTAACCATGTG........................................................................................... |
22 |
0 |
2 |
3.50 |
7 |
7 |
| ..................................................TTGTGCTTGATCTAACCATGTGT.......................................................................................... |
23 |
1 |
2 |
3.50 |
7 |
7 |
| ..................................................TTGTGCTTGATCTAACCATGTGG.......................................................................................... |
23 |
0 |
2 |
2.00 |
4 |
4 |
| ..................................................TTGTGCTTGATCTAACCATGTGGT......................................................................................... |
24 |
0 |
2 |
1.50 |
3 |
3 |
| ..................................................TTGTGCTTGATCTAACCATGTGAT......................................................................................... |
24 |
1 |
2 |
1.50 |
3 |
3 |
| ..................................................TTGTGCTTGATCTAACCATGTA........................................................................................... |
22 |
1 |
2 |
1.00 |
2 |
2 |
| ..................................................TTGAGCTTGATCTAACCATGTGG.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTAA.......................................................................................... |
23 |
2 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACTATGT............................................................................................ |
21 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTGTT......................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTGA.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTAGT......................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTAG.......................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGA............................................................................................ |
21 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTGGTAT....................................................................................... |
26 |
2 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTAGA......................................................................................... |
24 |
2 |
2 |
0.50 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAGCGATGTGA.......................................................................................... |
23 |
3 |
4 |
0.25 |
1 |
1 |
| ..................................................TTGTGCTTGATCTAACCATGTAAA......................................................................................... |
24 |
3 |
4 |
0.25 |
1 |
1 |