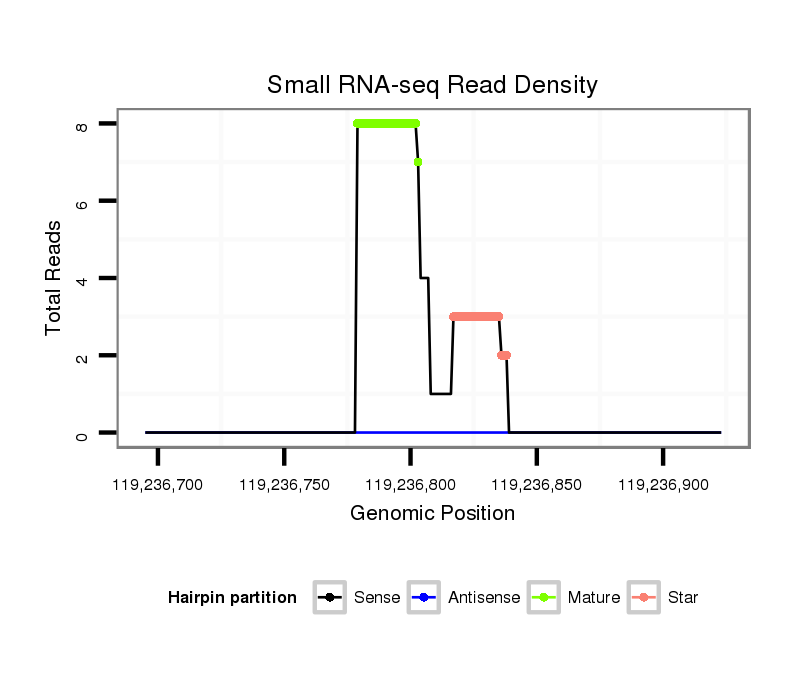
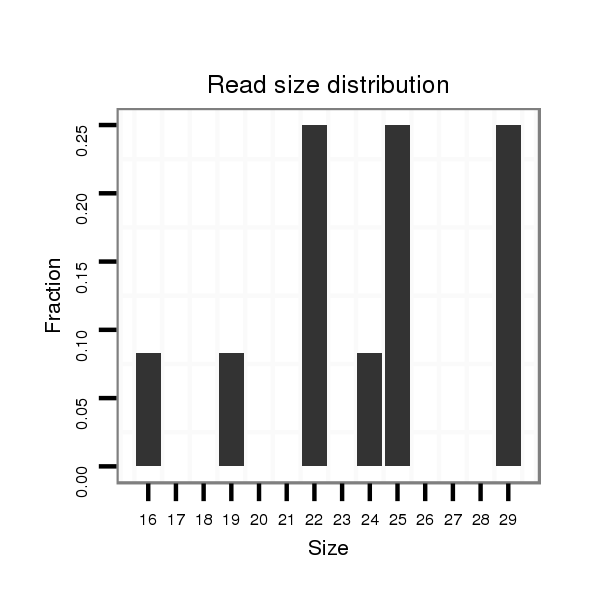
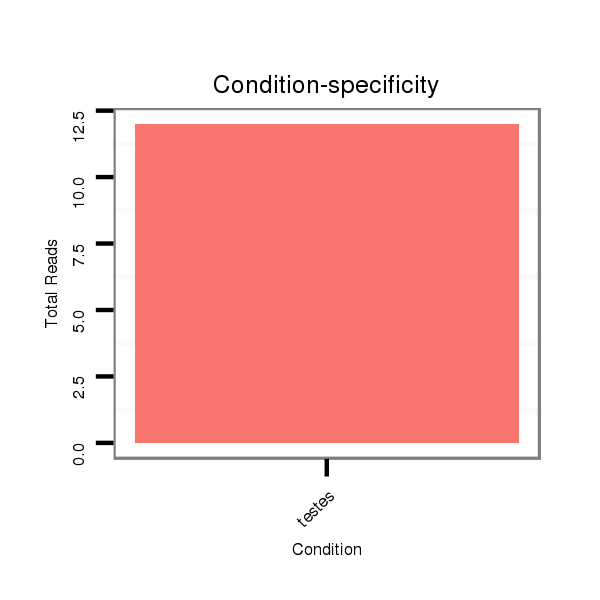
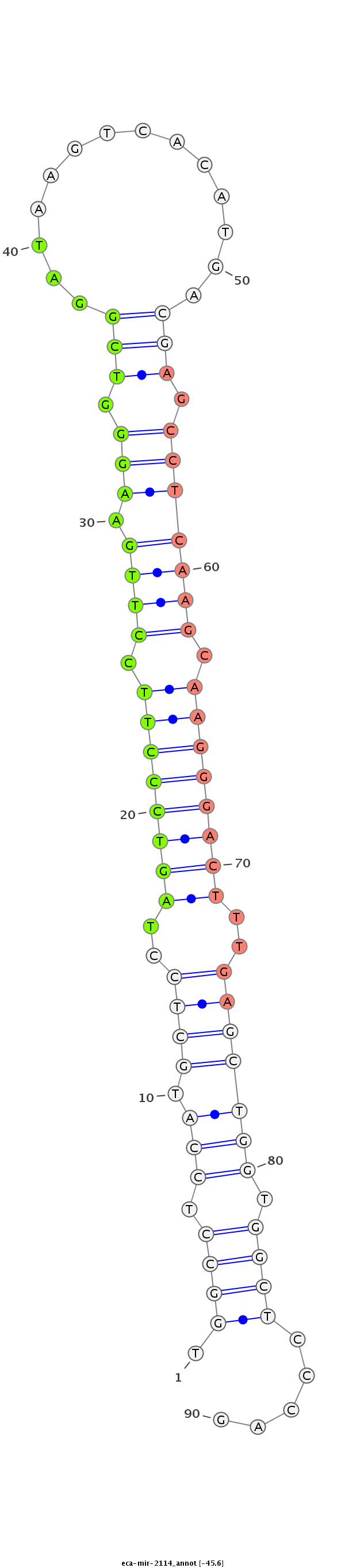
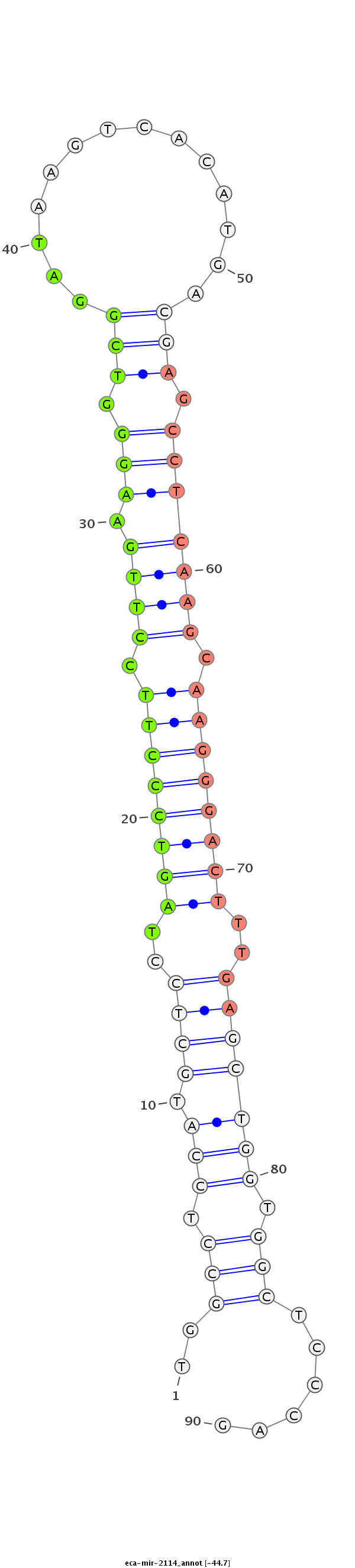
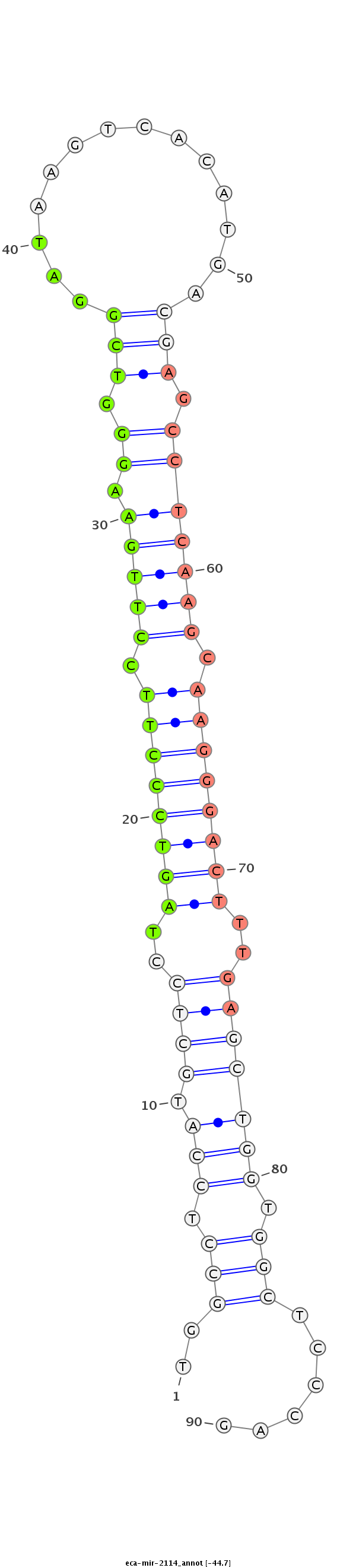
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGT..................................................................................... |
22 |
1 |
1 |
4.00 |
4 |
4 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGTA.................................................................................... |
23 |
2 |
1 |
4.00 |
4 |
4 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGAGT....................................................................................................................... |
26 |
2 |
1 |
3.00 |
3 |
3 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGAT........................................................................................................................ |
25 |
0 |
1 |
3.00 |
3 |
3 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGATAAGT.................................................................................................................... |
29 |
0 |
1 |
3.00 |
3 |
3 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGTAGA....................................................................................................................... |
26 |
2 |
1 |
2.00 |
2 |
2 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGATT....................................................................................................................... |
26 |
1 |
1 |
2.00 |
2 |
2 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGTT.................................................................................... |
23 |
2 |
1 |
2.00 |
2 |
2 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGA..................................................................................... |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ..........................................................................................................GATAAGTCACATGACG........................................................................................................... |
16 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGAAGACT.................................................................................................................... |
29 |
3 |
1 |
1.00 |
1 |
1 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGT.......................................................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCG........................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGGA......................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
| ....................................................................................TAGTCCCTTCCTTGAAGGGTCGAAC........................................................................................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGC..................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTT........................................................................................ |
19 |
0 |
1 |
1.00 |
1 |
1 |
| ..........................................................................................................................AGCCTCAAGCAAGGGACTTTGTAA................................................................................... |
24 |
3 |
2 |
0.50 |
1 |
1 |