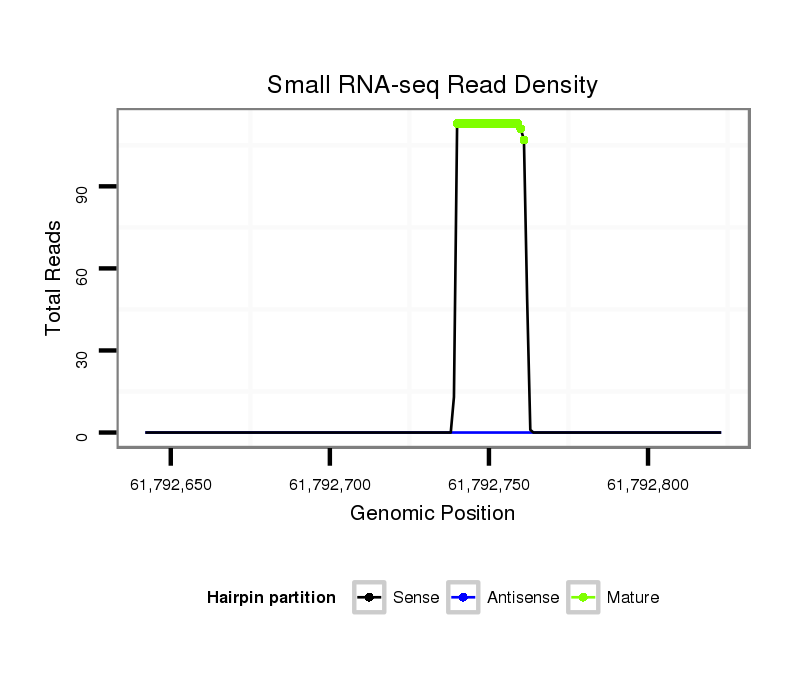
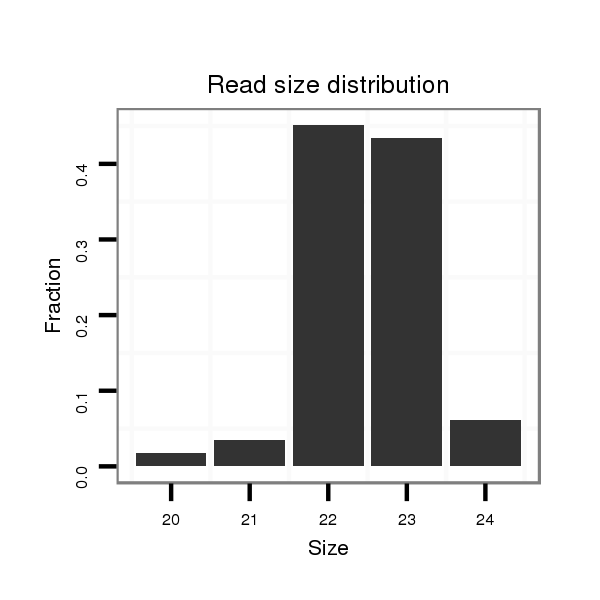
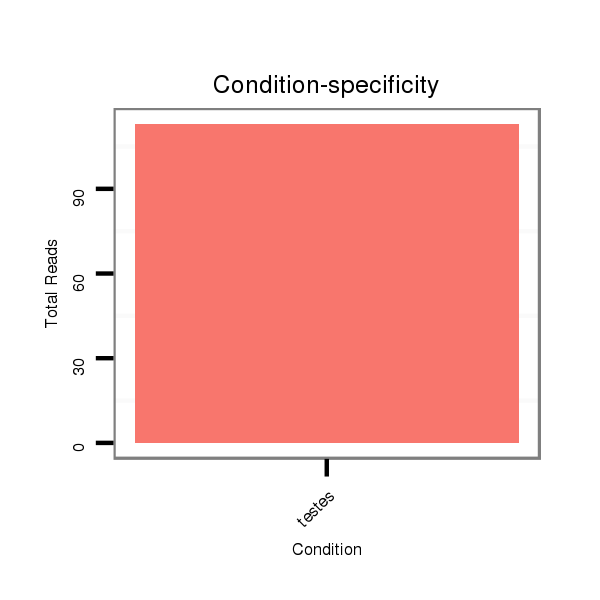
| ..................................................................................................TGTGCAAATCTATGCAAAACTG.............................................................. |
22 |
0 |
1 |
51.00 |
51 |
51 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTGA............................................................. |
23 |
0 |
1 |
42.00 |
42 |
42 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTGT............................................................. |
23 |
1 |
1 |
21.00 |
21 |
21 |
| .................................................................................................TTGTGCAAATCTATGCAAAACTG.............................................................. |
23 |
0 |
1 |
7.00 |
7 |
7 |
| .................................................................................................TTGTGCAAATCTATGCAAAACTGA............................................................. |
24 |
0 |
1 |
6.00 |
6 |
6 |
| ..................................................................................................TGTGCAAATCTATGCAAAACT............................................................... |
21 |
0 |
1 |
4.00 |
4 |
4 |
| .................................................................................................TTGTGCAAATCTATGCAAAACTGT............................................................. |
24 |
1 |
1 |
2.00 |
2 |
2 |
| ..................................................................................................TGTGCAAATCTATGCAAAAC................................................................ |
20 |
0 |
1 |
2.00 |
2 |
2 |
| ..................................................................................................TGTGCAAATCTATGCAGAACTG.............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCACATCTATGCAAAACTG.............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTGC............................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTGAT............................................................ |
24 |
0 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCGAAACTG.............................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAGAACTGT............................................................. |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGTAAATCTATGCAAAACTGA............................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTTA............................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .................................................................................................TTGTGCAAAACTATGCAAAACT............................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAAAAGTGT............................................................. |
23 |
2 |
2 |
0.50 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGCAAAACTGTA............................................................ |
24 |
2 |
2 |
0.50 |
1 |
1 |
| ..................................................................................................TGTGCAAATCAATGCAAAACTGA............................................................. |
23 |
1 |
3 |
0.33 |
1 |
1 |
| ..................................................................................................TGTGCAAATCGATGCAAAACTGT............................................................. |
23 |
2 |
4 |
0.25 |
1 |
1 |
| .................................................................................................TTGTGCAAATCTGTGCAGAACT............................................................... |
22 |
2 |
4 |
0.25 |
1 |
1 |
| ..................................................................................................TGTGCAAATCTATGGACAACTGT............................................................. |
23 |
3 |
6 |
0.17 |
1 |
1 |