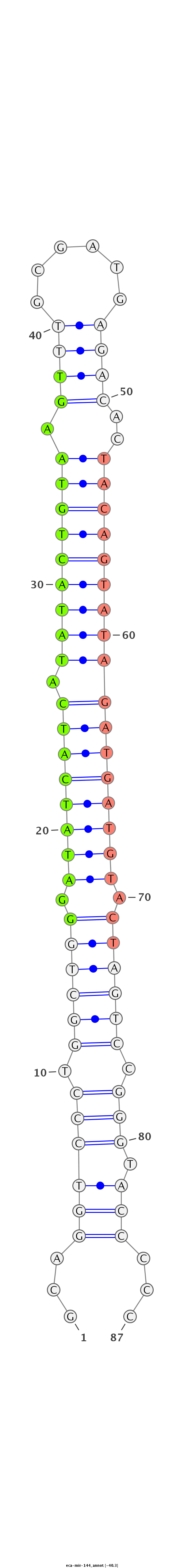
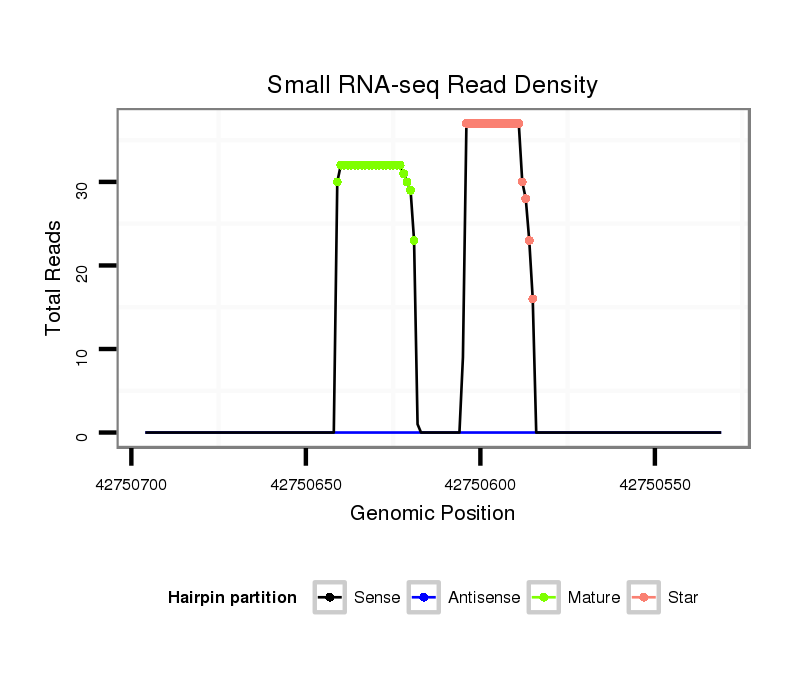
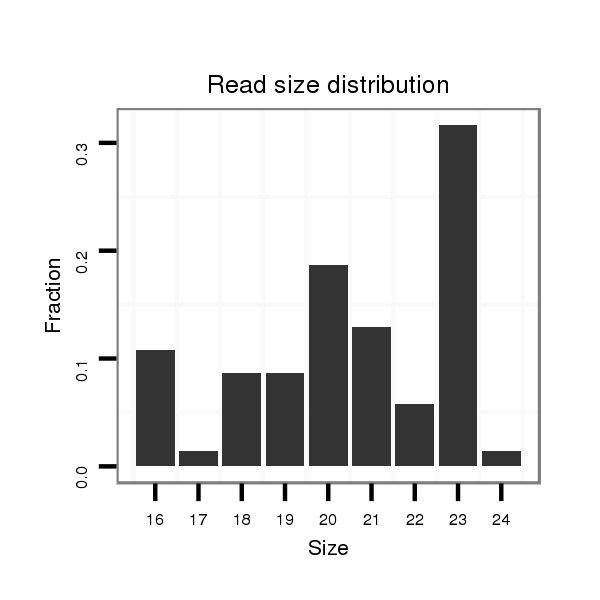
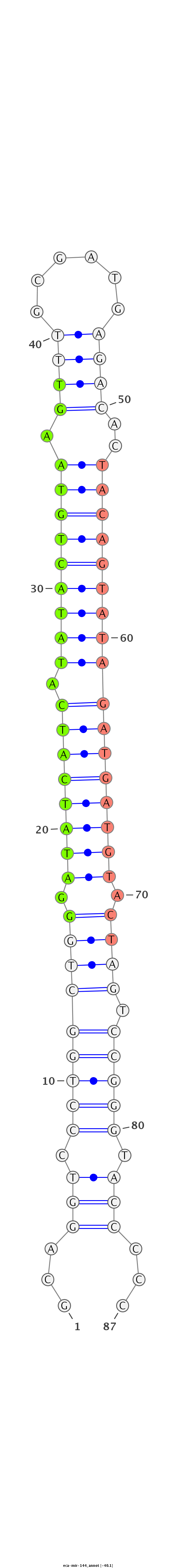
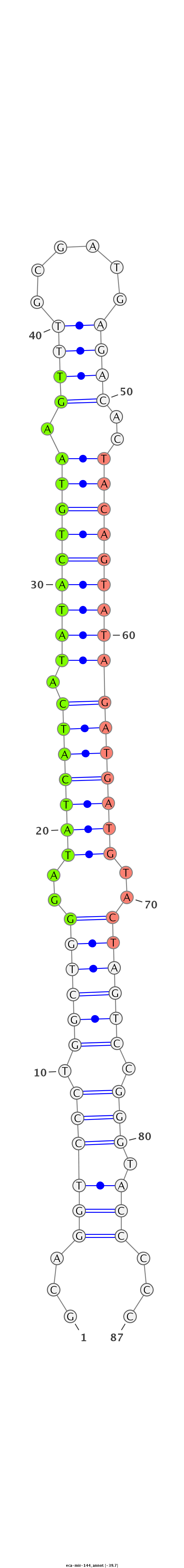
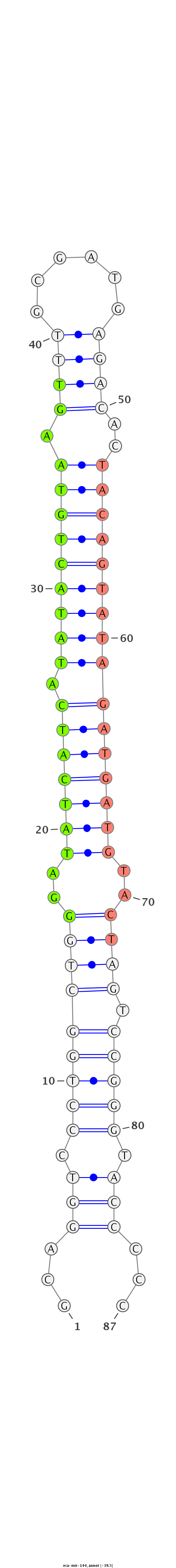
| .......................................................GGATATCATCATATACTGTAAGT........................................................................................ |
23 |
0 |
1 |
22.00 |
22 |
22 |
| ............................................................................................TACAGTATAGATGATGTACT...................................................... |
20 |
0 |
1 |
10.00 |
10 |
10 |
| ............................................................................................TACAGTATAGATGATG.......................................................... |
16 |
0 |
2 |
7.50 |
15 |
15 |
| ...........................................................................................CTACAGTATAGATGATGTACT...................................................... |
21 |
0 |
1 |
6.00 |
6 |
6 |
| ............................................................................................TACAGTATAGATGATGTA........................................................ |
18 |
0 |
1 |
5.00 |
5 |
5 |
| ............................................................................................TACAGTATAGATGATGTAC....................................................... |
19 |
0 |
1 |
5.00 |
5 |
5 |
| .......................................................GGATATCATCATATACTGTAAG......................................................................................... |
22 |
0 |
1 |
4.00 |
4 |
4 |
| ........................................................GATATCATCATATACTGTAAG......................................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
| ...........................................................................................CTACAGTATAGATGATGTAC....................................................... |
20 |
0 |
1 |
2.00 |
2 |
2 |
| .......................................................AGATATCATCATATACTGTAAGT........................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATACTGT............................................................................................ |
19 |
0 |
1 |
1.00 |
1 |
1 |
| ............................................................................................TACAGTGTAGATGATGTACT...................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................................................................TACAGTATAGATGATGT......................................................... |
17 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATGTACTGTAAGT........................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATGCGGTAAGT........................................................................................ |
23 |
2 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATACTGTAAGTT....................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATACTGTAA.......................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................CTACAGTATAGATGATGT......................................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATACTGTTAGT........................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................................................................TACAGTATAGATGATGTACG...................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................GGATATCATCATATACTGTA........................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |