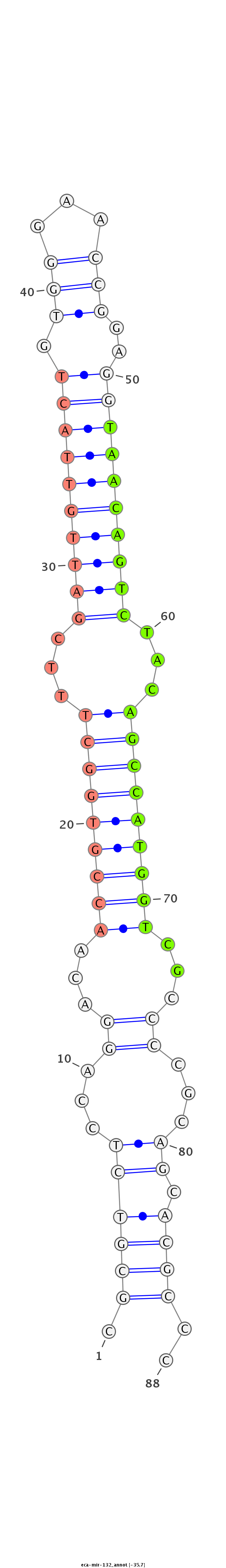
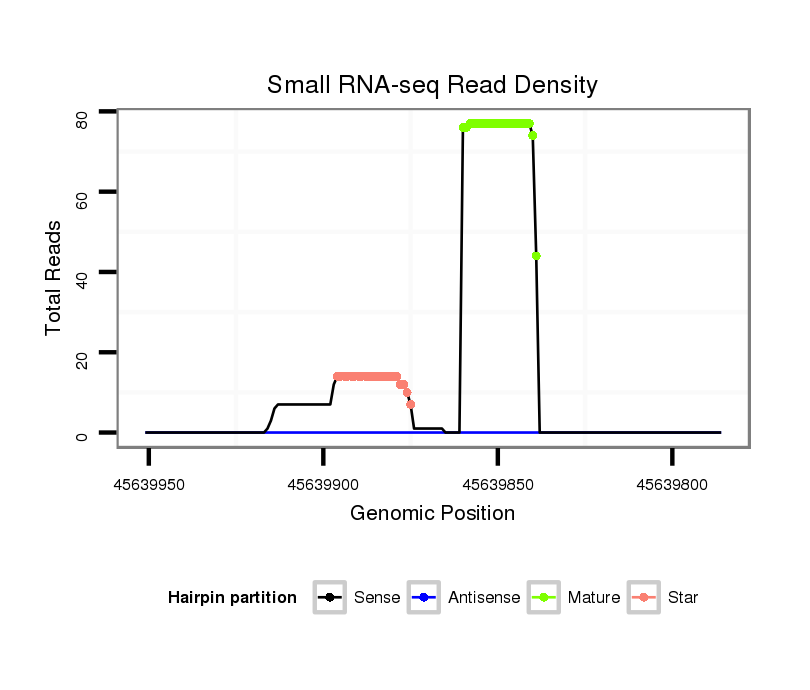
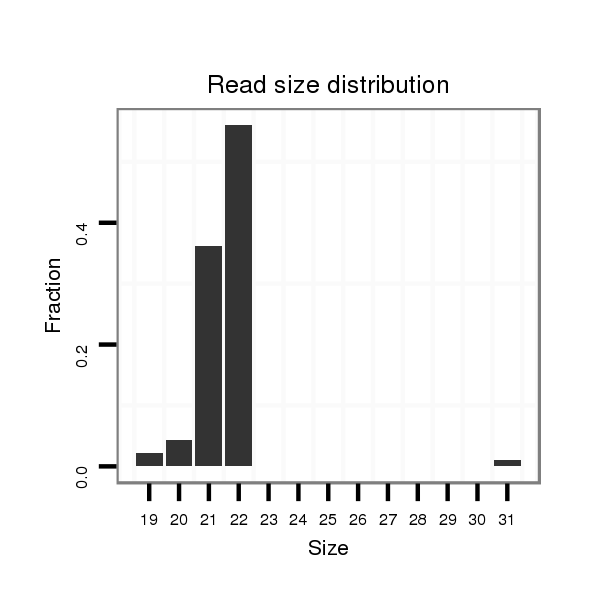
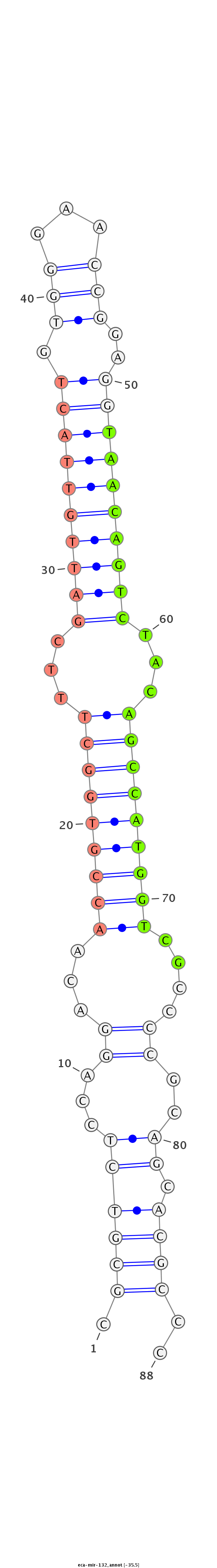
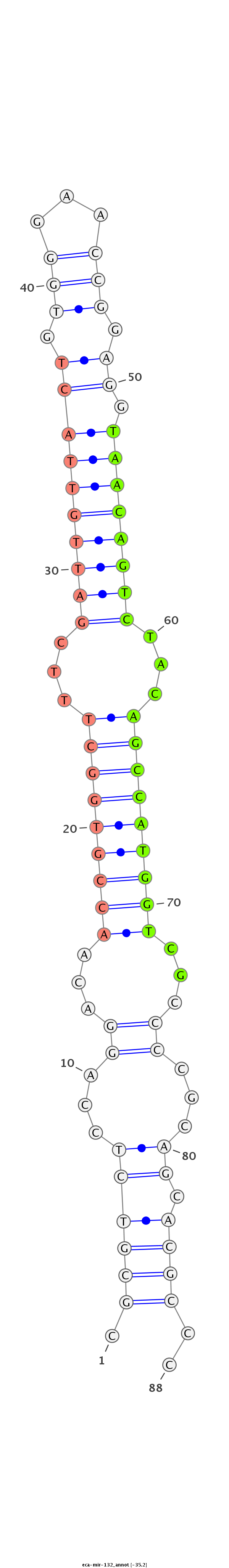
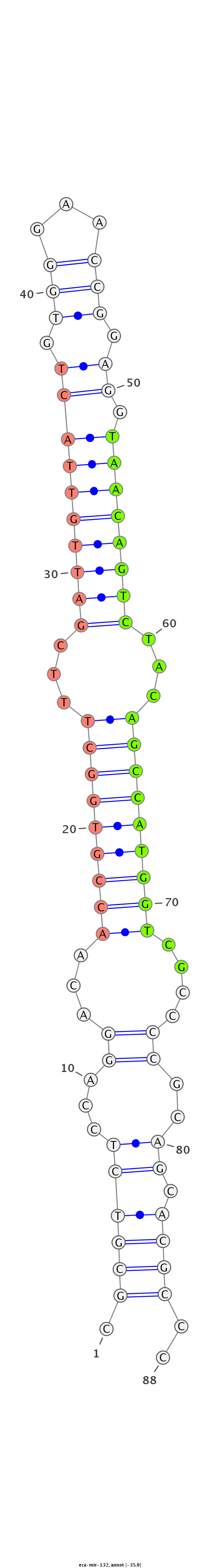
| ...........................................................................................TAACAGTCTACAGCCATGGTCGT.................................................... |
23 |
1 |
1 |
44.00 |
44 |
44 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCG..................................................... |
22 |
0 |
1 |
43.00 |
43 |
43 |
| ...........................................................................................TAACAGTCTACAGCCATGGTC...................................................... |
21 |
0 |
1 |
30.00 |
30 |
30 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCT..................................................... |
22 |
1 |
1 |
9.00 |
9 |
9 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCC..................................................... |
22 |
1 |
1 |
7.00 |
7 |
7 |
| .......................................................ACCGTGGCTTTCGATTGTTACT......................................................................................... |
22 |
0 |
1 |
6.00 |
6 |
6 |
| .....................................CCCCGCGTCTCCAGGACA............................................................................................................... |
18 |
0 |
1 |
3.00 |
3 |
3 |
| ...........................................................................................TAACAGTCTACAGCCATGGT....................................................... |
20 |
0 |
1 |
3.00 |
3 |
3 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGTT................................................... |
24 |
2 |
1 |
3.00 |
3 |
3 |
| ......................................................AACCGTGGCTTTCGATTGT............................................................................................. |
19 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................AACCGTGGCTTTCGATTGTTAC.......................................................................................... |
22 |
0 |
1 |
2.00 |
2 |
2 |
| ......................................................AACCGTGGCTTTCGATTGTTA........................................................................................... |
21 |
0 |
1 |
2.00 |
2 |
2 |
| ............................................................................................AACAGTCTACAGCCATGGTCGT.................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
| ....................................CCCCCGCGTCTCCAGGACA............................................................................................................... |
19 |
0 |
1 |
2.00 |
2 |
2 |
| ...........................................................................................TAACAGTCTACAGCCATGGGCT..................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
| ......................................CCCGCGTCTCCAGGACA............................................................................................................... |
17 |
0 |
2 |
1.00 |
2 |
2 |
| ...........................................................................................TAACAGTCTACAGCCAGGGTC...................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| .......................................................ACCGTGGCTTTCGATTGTTAA.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| ............................................................................................CACAGTCTACAGCCATGGTCGC.................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
| .............................................................................................ACAGTCTACAGCCATGGTCG..................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAACCATGGTC...................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGATT.................................................. |
25 |
3 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGAAC.................................................. |
25 |
2 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTAAAGCCATGGTCC..................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
| ...................................GCCCCCGCGTCTCCAGGAC................................................................................................................ |
19 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................ACCGTGGCTTTCGATTGTTACTGTGGGAACC................................................................................ |
31 |
0 |
1 |
1.00 |
1 |
1 |
| .......................................................ACCGTGGCTTTCGATTGTTACTGTGGGAACT................................................................................ |
31 |
1 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCAAGGTCGT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCAT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| .......................................................ACCGTGGCTTTCGATTGTTAC.......................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGTACT................................................. |
26 |
3 |
1 |
1.00 |
1 |
1 |
| .....................................................................................CCGTGGTAACAGTCTACAGCCATGGTCGC.................................................... |
29 |
2 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCTT.................................................... |
23 |
2 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGTAC.................................................. |
25 |
2 |
1 |
1.00 |
1 |
1 |
| ...........................................................................................TAACAGTCTACAGCCATGGTCGG.................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
| .............................................................................................ACAGTCTACAGCCATGGTCGT.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
| .............................................................................................ACAGTCTACAGCCATGGTCGTTT.................................................. |
23 |
3 |
2 |
0.50 |
1 |
1 |
| .............................................................................................CCAGTCTACAGCCATGGTCGTA................................................... |
22 |
3 |
3 |
0.33 |
1 |
1 |