| droYak3 |
3R:945790-946040 - |
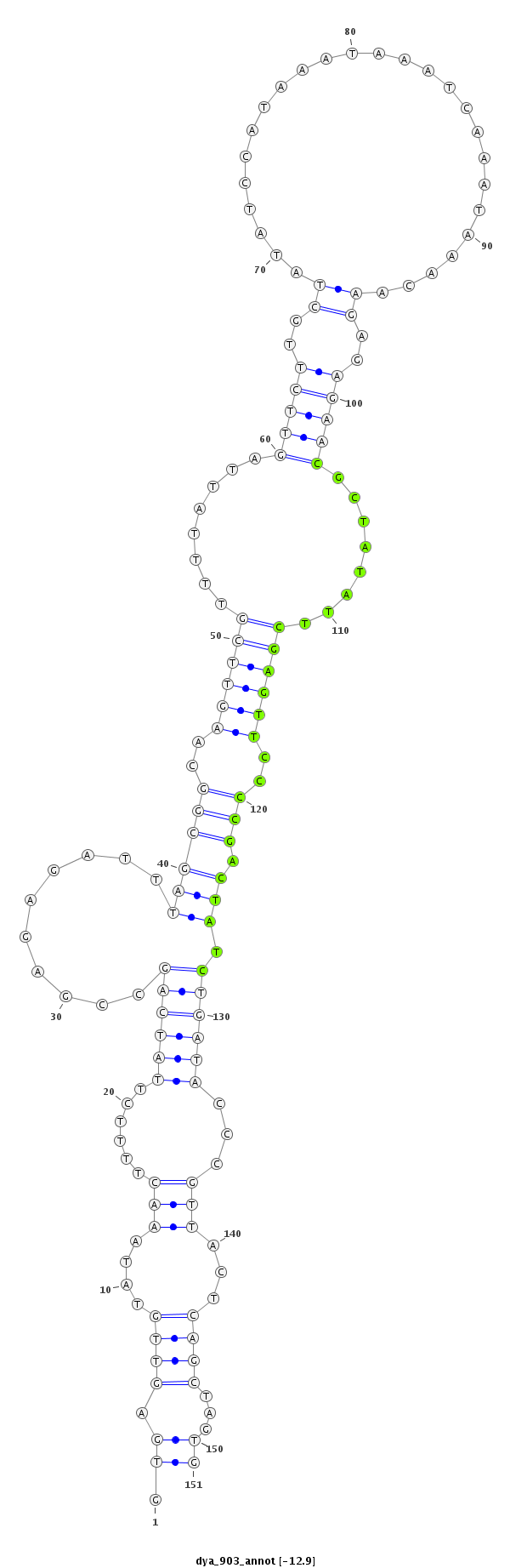
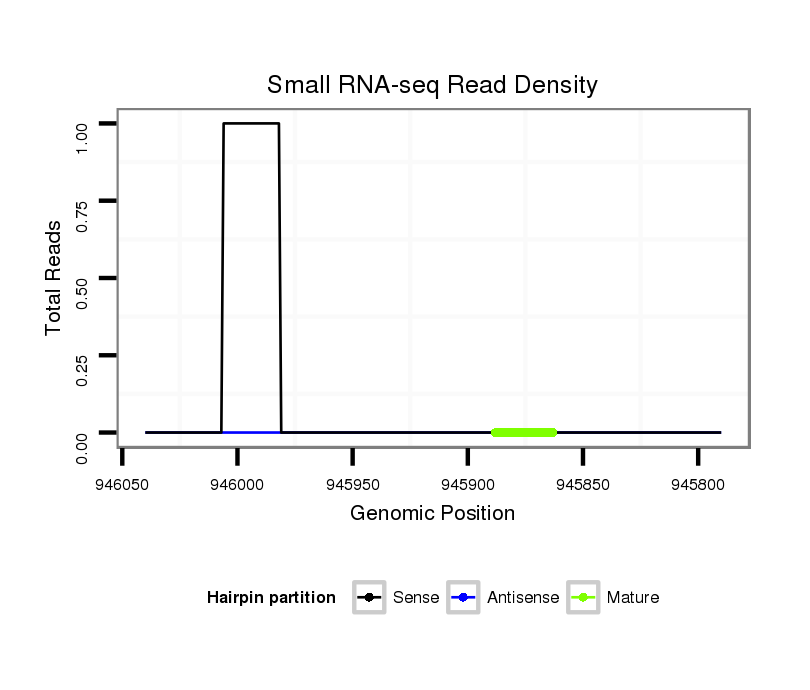
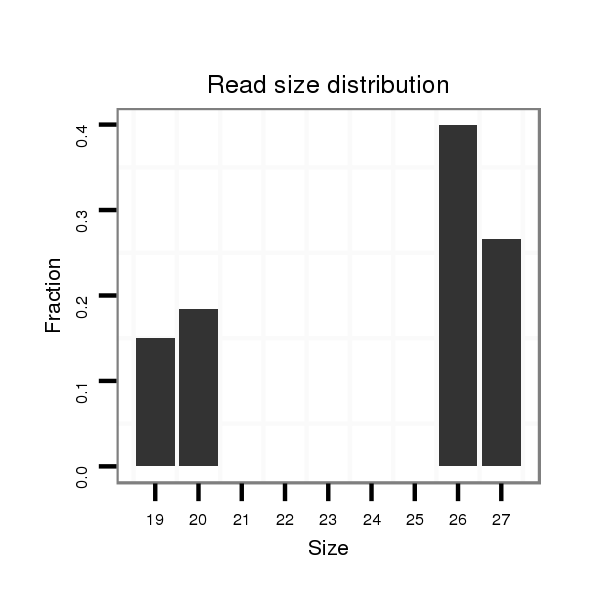
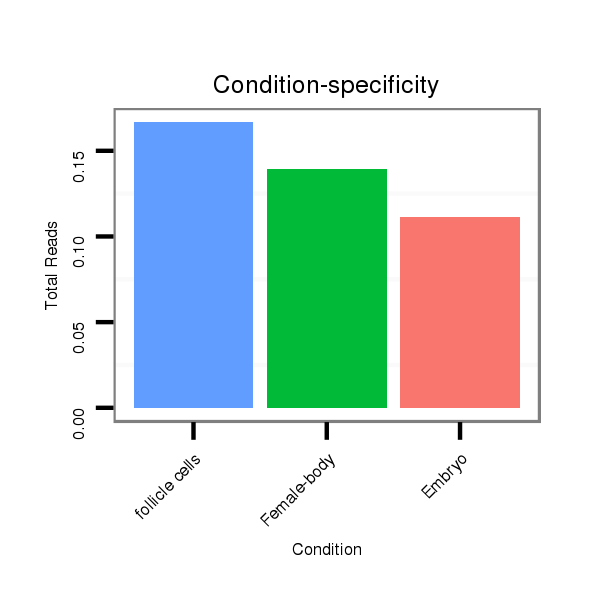
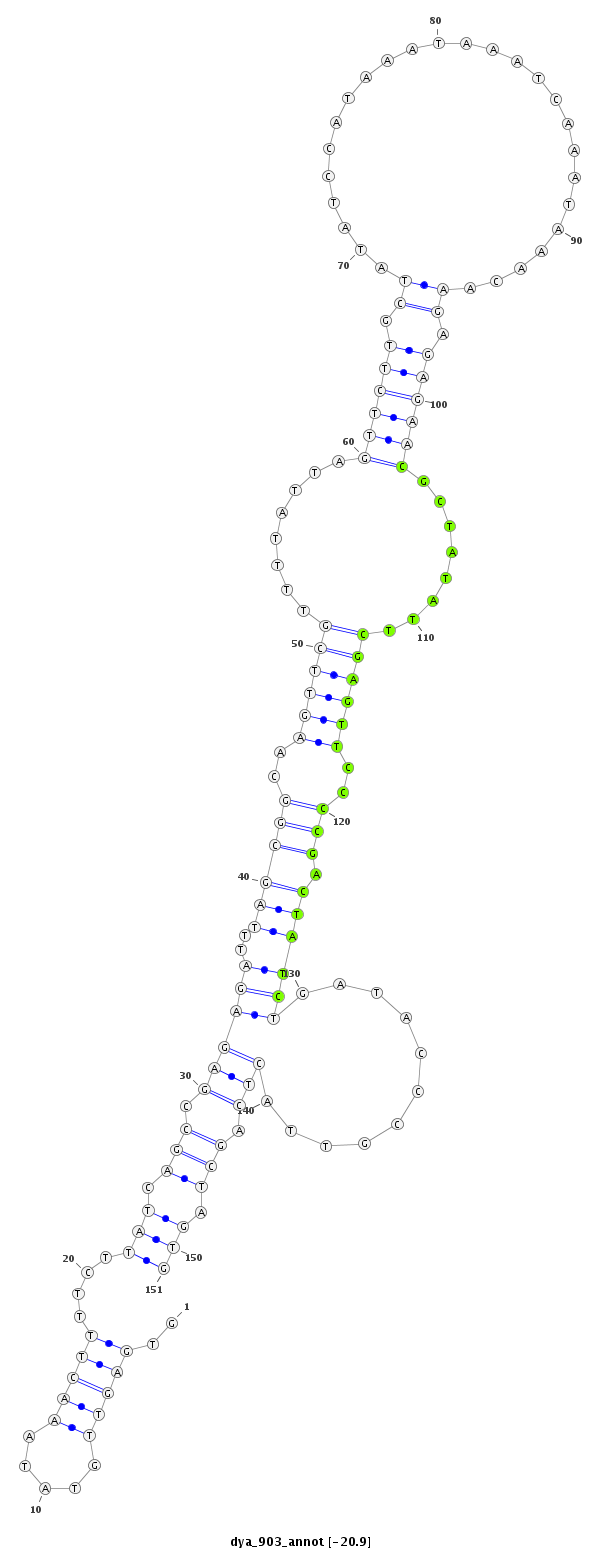
dya_903 |
TAACC-AGC-----------ACAACTGCGGCAG--CAACAAAGCTAGCATCAGTAGCCACCAATGTGAGTTGTATAAACTTTT---------CTTATCAGCCGAGAGATTTAGCG---------------GCAAGTTCGTTTTATT---------------------------------------------------------------------------------------------------------------------AGTTCTT---------------------------------------------------------------------------GCTATATCCATAAAT--------------------------------------------------------------------------------------------------------------------------AAAT----CAAAT----AAACAAGAGAGAAC-GCTATATTCG--AGTTCCCCGACTATCTGATACCCGTTA----CTCAGCTAGT------------------------------------GGAAGGGCGAAGGAGAGT----------CTTAAACA--------------------------------------------------CAGTT----------------------------------TTTGACGGTTTGTAGGCGTT |
| droEre2 |
scaffold_4929:18000465-18000699 - |
|
GCAGC-AGC-----------GCAGCTTCGACAA--GGGCGACTCTG--------------------------------------------------------------------------------------------------------------------------------GCATTGA-GAACGATTTCCGGAAGGAGTCCTTCAACGGGGACT-------------------TGAC--C-ACAAGGTAGGC--------GAATAATCTTGCCTT---------------------------ATATAGGTATCAA------------T-------------------------------------------------------------------------------------------------------------------------------------------------------TTGT----AA----TAACTTTATGTGAAAAC-AGAAGAGTAG--AGTTTCTCGACTATCAGATACCCGTTA----CTCAGCTAGT------------------------------------GGAAGTGCGAACGAGAAA----------TTTCA----------------------------------------------------------------------------------------GCTTTCTTACGCCA-TTTCGATAG |
| droSec2 |
scaffold_30:518956-519175 + |
|
TGTGATT-TAT----------------T-----AC----------------------------------------------T-------------ATCGTGTGATGGATCTGACG---------------TGACGGTATCA-------------------------------------------------------------------------------------------------------------------------------------CTAT---------------------------CAGTATTT--------------------------------GCCAGATCTGTAAAG--------------------------------------------------------------------AG-------------AGAAAT---------------------------------GTTT----ACATT----AAACAAGAGAGAAT-GCTATAATCG--AGTTTCCCGACTATCTGATACCCGTTA----CTCAGCTAGTGT-----------------------------------GTATTGGGGAATAGAT-TAAGAAAAAATTTAAAAATTAAACGTCA----------------------------------------CCGG-----------------------------------TTCGGAGGCCTTAGGGCGTT |
| droSim2 |
3r:394850-395031 + |
|
TTGCA-C-AAA-------------------------------------------------------------------GACTC---------GTTATCA------------------------------------------------------------------------------GTGG-CG----------------------------------------------------------------------------------------------------------------------------------------------ACAATTTGTGCGAGTGCCTAATAATAACAGAAAG-------------------------------------------------------------------------------------A------------------------------------ATCC----TAATC----AAACAAG-GGGATC-GCTATAGTCC--AATTTCTCGAATATCAGATACCCGTTA----CTCAGCTAGT------------------------------------GGGATTGCGAACAAAATA----------TTTGTTTC--------------------------------------------------AAATC----------------------------------GATAGAAGTTTGTAAGCGTT |
| dm3 |
chr3LHet:2255683-2255857 - |
|
TATTA-A-CATTTCCAGAAAAGAAATAT-----------------------------------------------------AA---------CAGTTC-----AAG-----------------------------GGC-----CTT---------------------------------------------------------------------------------------------------------------------TACTAGTG------------------------------------CTAAATATCCG------------G-------------------------------------------------------------------------------------------------------------------------------------------------------TTAT----AAATA----AAACAAAAGAAAAT-GCTATAGTCG--AGTTCCCCGACT-CGAGTTAGCCGTTA----CTTAGCTAGG------------------------------------GGA-ATGCGAACACATTG----------A---------------------------------------------------------CCGTT----------------------------------TTTGGTGTTTTGTAGGTGAA |
| droEug1 |
scf7180000407642:33735-34047 - |
|
AACTA-G-CAC-------------------------------------------------------------------CGCA-------------AT----------------------------------------TTCTTTGACAACGTCAACCATAACAACAACAACAACCGCAACAG-TAACG-------------------------------ACAGCGCGATTAACGACGTCAT------------------------------------------------------------------------------------------------------------------------CAGCTAGCAGCGAAAACATTA------------------------------------------------------------------------ATACTTCTG-CTGCTGCGCTACCCGAAAAAGAG----AA----CAAAAACAAGAGGAAAC-GCTATAGTCG--AGTGCCGCGACCATAAGATACCCGTTA----CTGTTTTACA----------AAATTTTAAAATATTATTTTGCTAGTGGAAATTCACAAGG---------------------------CGCCACCTATTGTCAGAGGAAGTATGTTGAGTATGCTTATAGTGAC-GTCAC----------------------------------TGCCCAT-GTGAGGCCTT |
| droBia1 |
scf7180000302155:2883100-2883306 + |
|
GACAA-C-AAA-------------------------------------------------------------------AATTCTAGTTTCGT-----------------TATCCTTTAACCTGGGGTAC---------------------------------------------AGATCACTTTTCG-------------------------------AT---------------TTCAGACTG---------TTGAATATAA--------------TTTGTTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTCGGA-TTGTTATATTG--------TTAA----AA----AAAAAACAA------------GCAATCG--AGTGCCTTCACTTCGAGACACCCGTTA----CTCAGCTAAC------------------------------------AAACTTAAAAATA-----------------------------------------------------------------------------------------------------TCCCATTGATATTTAGTCTGTTGTTGGCGTT |
| droTak1 |
scf7180000414323:115802-116075 - |
|
TAACA-A-AAA----------CAAAAGCAACAG--CA-----------------------------------------------------------------------------------------------------------------------------------------ACAAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCTGGCACCGGTGGCATTGCCGTAGTTTTGGCATCAACATAATTACTTGTTTCCACTTTTGTACTCGGCTTCCACTGTCGCAGTTGCGTTTTTATTGTGGCCCACT----------------GTATC-TAAT--TAAAAACTAGAGAGAAC-GCTATAGTCG--GGTGCCCCGACTATCAGATACCCGTTA----CTCAGCTAAA------------------------------------GGGAGTGCGAAAGAGAT-GGAGAAAA-----------------------------------------------------------AC--------------------TTTGATCCGCCGTAACTTTTTAACGAATGGTCCGATTT |
| droEle1 |
scf7180000490632:10605-10818 - |
|
AGTTG-T-ATC--------A--C-------------------------------------------------------TTT--------------ATCTTG-----------------------TAGAC---------------------------------------------GGAACAT-TCACG-------------------------------AC---------------ATTGCGCCG---------GTAAATTTAT--------------TTTGTGCTTGCCATAAAA----------------------CCCCTTCAAAAAACTTTTG-------------------------------------------------------------------------------------------------------------------------------------CAATT--------CCAG----AA----TAAAAACAAGAGAGAAC-GCTATAGTCG--AGTTTCCCGACTATTAGATA------A----CTCAGCCAAC------------------------------------AAA-CTTCAAACAGAGCAGAGGA-----C-------------------------------------------------------------------------------TTAATAACCCTTAA----------CTTCAAAAATATA |
| droRho1 |
scf7180000780072:113201-113472 + |
|
TAAAT-CGG-----------ACAATTGTAACTA--TATTAAAATAATCATCAAAAATCAATAAAGAAATCAATATAATCT-----------------------------------TTTGCGTGGCATCCAGCAAATTTGTAT------------------------------------------------------------------------------------------------------GCATAGAAT---------------------------------GCGATAAAAACTCGCAAATATGTCT---------------GAAACTTGTG-------------------------------------------------------------------------------------------------------------------------------------CA-------------------GT----GAAAAACAAGAGAGAAC-GCTATAGTCG--AGTGCCCCGACTATCAGATACCCGTTA----CTCAGCTAAA------------------------------------GGGAGTGCGAGGGAGAT-GGAGA---------------------------------------------------------TAGAAAC--------------------TTTGATCACGCATAACTTTTTAACGAATGGTCCGATTT |
| droFic1 |
scf7180000450041:9702-9881 - |
|
AATTT-C-AAT----------------------------------------------------------------------------------------------------------------------------------CTGTTAACTTGA-----------------------------------------------------------------------------------------------------------------ACT------------------------------------AACCGCATACAAC-----------------------------------------T--------------------------------------------------------------------------------------------------------------------------CTG---CCAAAAC----AAACAAGTGAGAAC-GCTATAGTTG--GGTGCCCCGACTATCAGTTACCAGTTAAATACAGAGCCAGA------------------------------------TGTATTGTAT-TGTGA------------TTTAAATA--------------------------------------------------AAGTTCCTGCATTGGCTTAGA----AGAG----------TTCATTGGCTTATGAGCA-T |
| droKik1 |
scf7180000302682:1553037-1553138 - |
|
AAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAAAGAAAGAAC-GCCATTGTCG--AGTGCCCCGAATATGAGATACCCGGCA----CTCAAG---GCTCTATTACAAAACA-------------------------------------T-TAAACAAAGACTCAAATA--------------------------------------------------CAGCT-----------------------------------------------------T |
| droAna3 |
scaffold_12947:768524-768658 + |
|
CTTAT-T-AAT----------------C-----AA----------------------------------------------GA---------CATTTT-----AGA---------------------------------------GACCTC----------------------------------------------------------------------------------------------------------------AA-----------------------------------------------------------------ATTGTATTACTT---GTTAGTCCCCTAACA--------------------------------------------------------------------------------------------------------------------------AAGTCCCCAAAACAAAAAAAAAACAAAGAACGGCTGAAGTCG--AGTTTCCCGACTGTATAATATCCGGTA----TTCATCTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396426:362291-362423 - |
|
TCTCCTC-TAT----------------C-----GAG------------------------------------------AGCTTCAGCTTCGC-----------------TAT-------------------------------GTC---------------------------------------------------------------------------------------------------------------------TAGTCGTGCCGCACT---------------------------CTCTGTTTATTAC-----------------------------------------A--------------------------------------------------------------------------------------------------------------------------ATAC----ACAAT----AAATTAGAAAGAACGGCTATATTCG--AGTTCCCCGACTATTTTATACACGGTA----CTCGGTAAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp5 |
3:9376153-9376246 + |
|
ATTCA-G-TTG-------------------------------------------------------------------CACT-------------GT----------------------------------------CGTTCCATTAATGAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACT---------------------------------------------------------------------------------------------------------------------------------------ACA----A--------CAACAAACTAAAGA-GATGTAGTCG--AGTTTTCCGACTATTAAATACCCAGTA----CTCGG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droPer2 |
scaffold_6:456028-456138 - |
|
GGAAA-G-TTC--------G--T----T--------------------------------------------------------TAC--------ATCTTT---------TA-------------------------------ATT---------------------------------------------------------------------------------------------------------------------TATTTGTG------------------------------------CTATTTTTAA------------------------------------------C--------------------------------------------------------------------------------------------------------------------------ATTT----TTTTC----AAACGAGAAAAAAT-GCCTTAGTCG--AGGGTTCCGACTACAAAATACCCGCTT----CTCAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004914:14624-14754 + |
|
ATATT-A-ATA--------A--ATAAATAAATA--TA-----------------------------------------------------------------------------------------------------------------------------------------GTAACGA-TCGCT-------------------------------------AAATCAACG---------CT---------ACATA-----TATATATTCTGCGC------------------------------------------------------------------------GCATACTAAGC--------------------------------------------------------------------------------------------------------------------------CAGTATTCATA---CTAAAATCAGAGAGAAA----------------------------------------------------------------------------------------------------------------------------AT--------------------------------------------------CAGTT----------------------------------TTGTTTAATTTGCGGGGGCG |
| droVir3 |
scaffold_12734:344626-344700 - |
|
TCTTA-T-TCC--------A--A----C-----A--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCAAAT--------------------------------------------------------------------------------------------------------------------------AAAT----TAATA----AAACAAGTAAGAGT---TCTAGTCGGGAAGCTCCCGACTAGGGGATACCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6500:8323985-8324173 + |
|
ACTGC-AGCAG---------GTGGCAACGAAAA--CAGCGCTTATT--------------------------------------------------------------------------------------------------------------------------------GTAATAA-GCACGAGTCAAAGTGCAATTTCTTTACAAACTTCCCAC---------------AGTACATGG---------ATAGATTAAG--------------AGGGATGTT--------------------------------------AGAA--------TAGTTT---GTCAATTCCTTAAAA--------------------------------------------------------------------------------------------------------------------------AAAT----CTATT----AAACAAGTAAGAAG-GCTCTAGTCGA-GACTGCTCGACTAGGAGATACCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15110:14049376-14049517 + |
|
TTGTT-T-ACC--------A--A----C-----AAGAAGTTTTGCG--------------------------------------------------------------------------------------------------------------------------------GCAATGG-ATGTGGTTTGTGGTTTAACTGCTTTTATGGTAACA---------------------------------------------------------CAAA--------------------------TATATATATATATA-----------------------------------------T--------------------------------------------------------------------------------------------------------------------------ATAC----AA----TAAAAACAAGTAAGAAC-AACTGCTTCA--GGCTCCTCGAACATTACATACCC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |