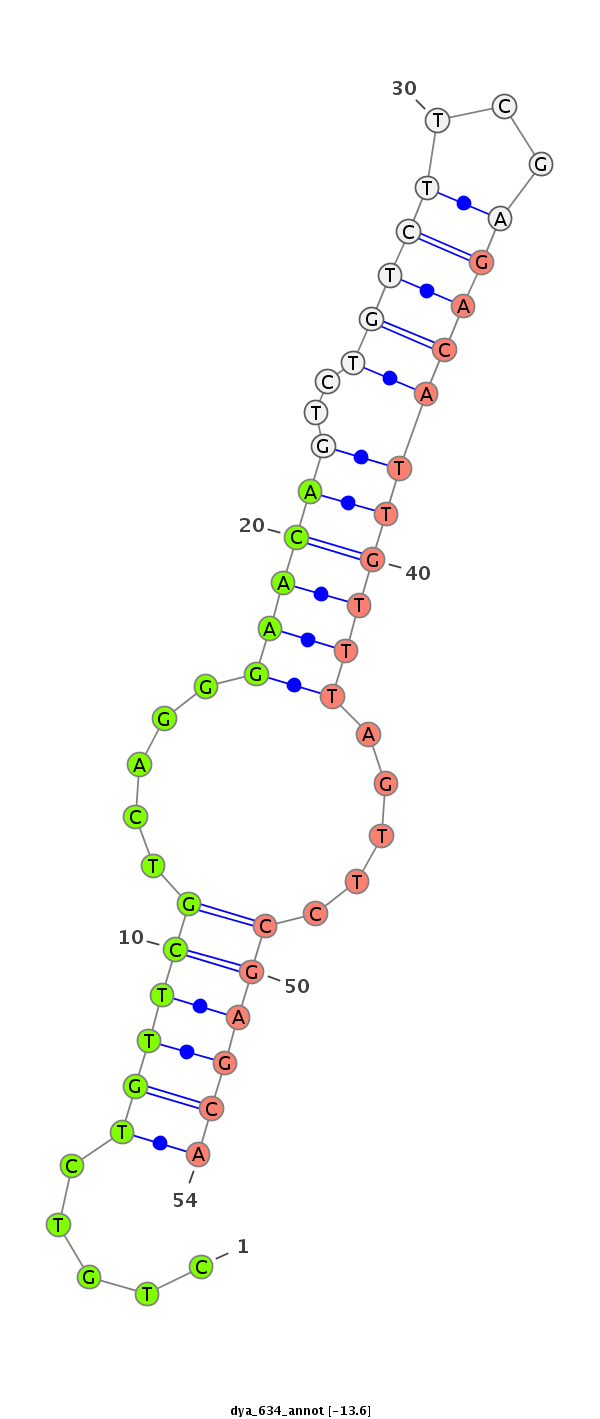
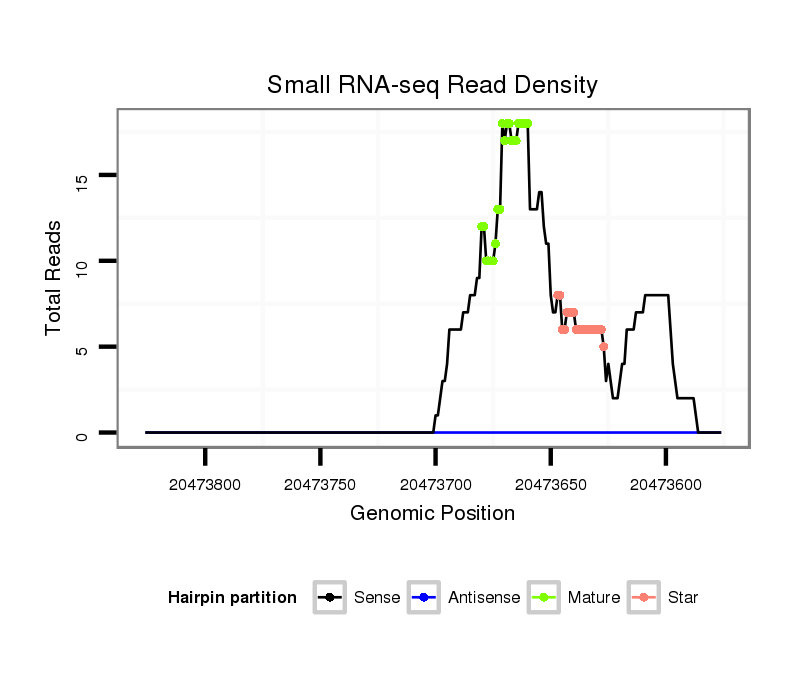
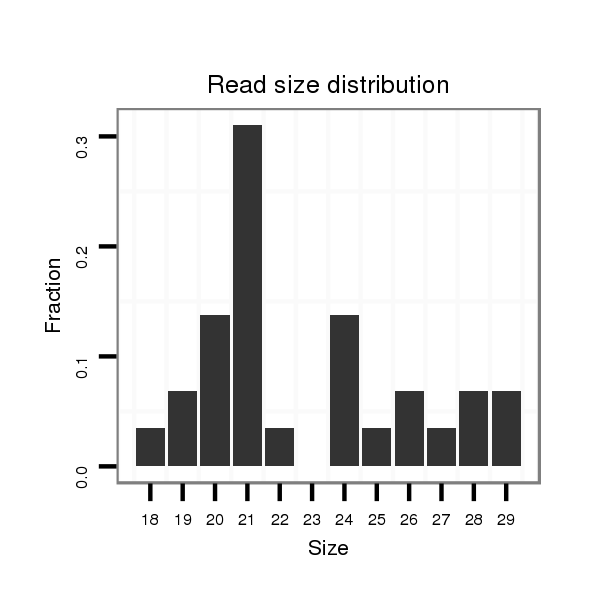
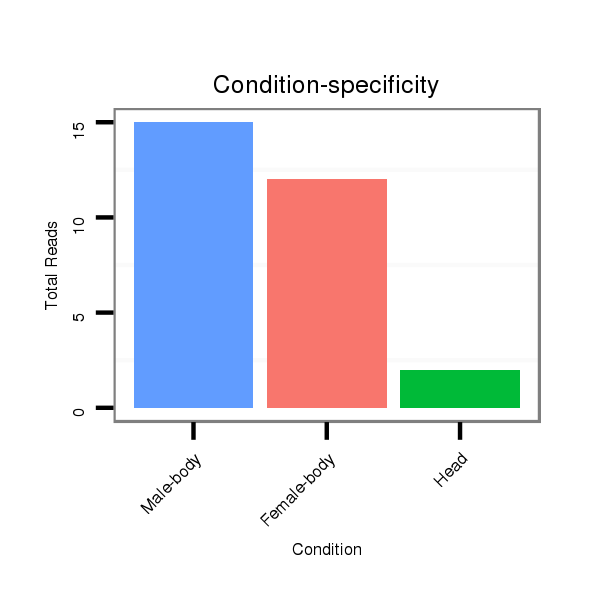
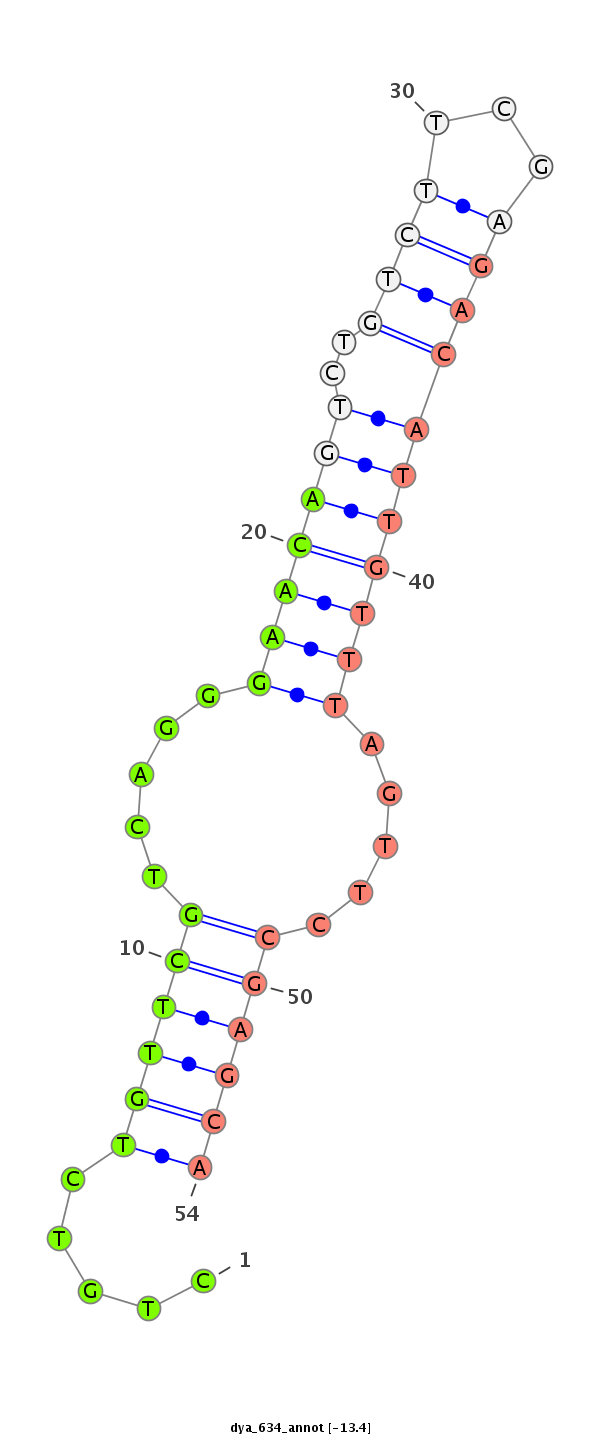
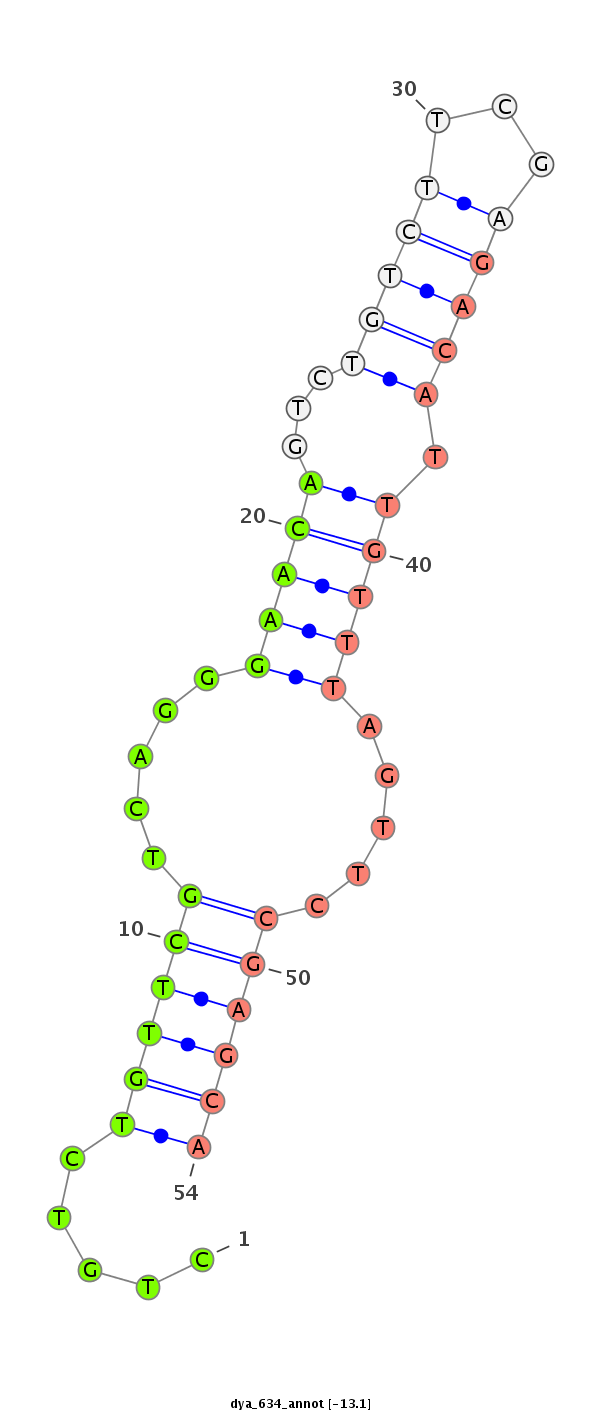
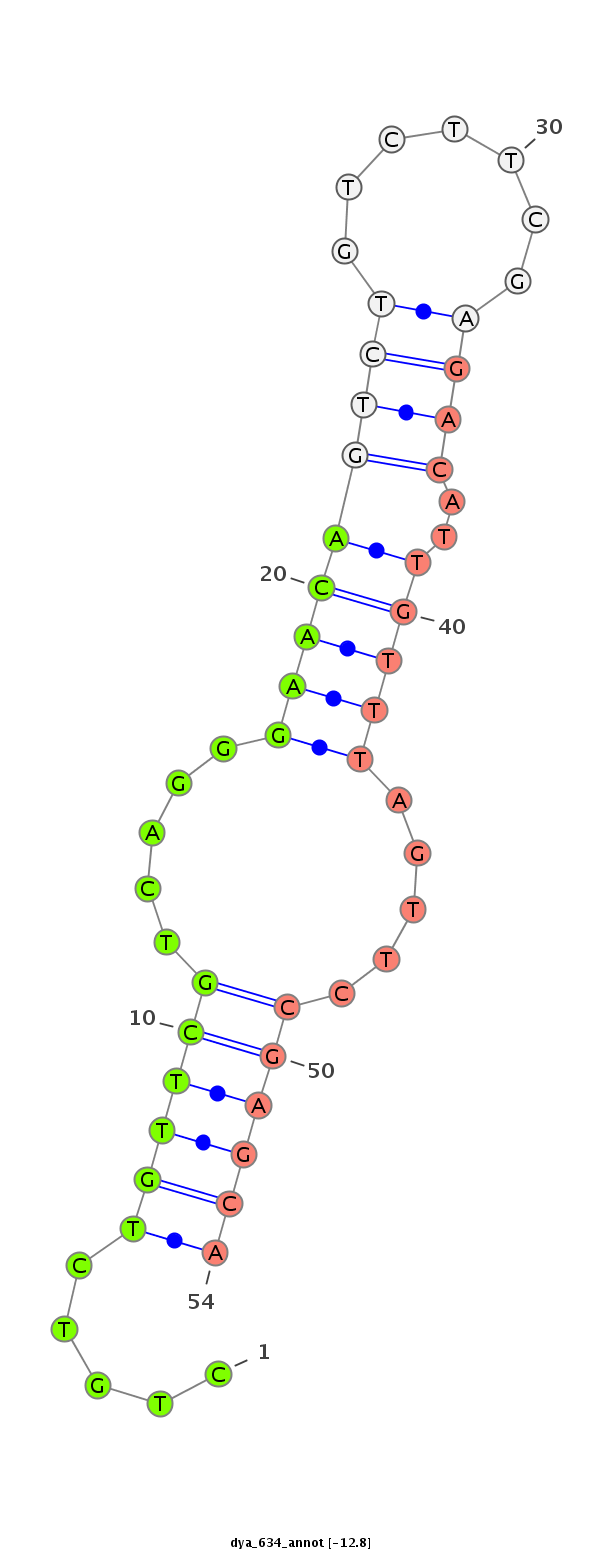
ID:dya_634 |
Coordinate:2R:20473626-20473776 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
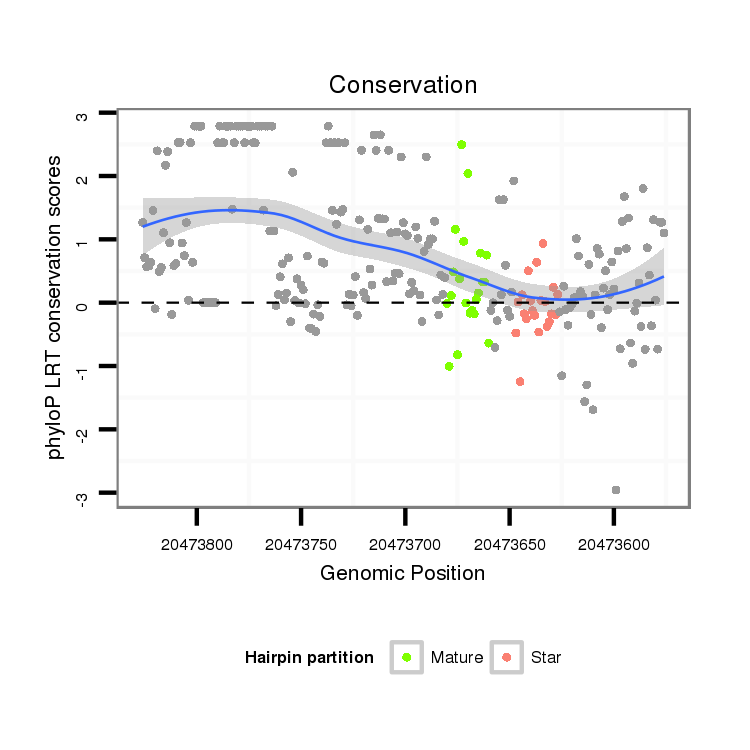
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.4 | -13.1 | -12.8 |
 |
 |
 |
exon [dyak_GLEANR_11765:3]; CDS [Dyak\GE11433-cds]; intron [Dyak\GE11433-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CTGGCTCGACACTGGTTTTGTCGCCTTATCAGCTATCAGCTAATGATTACAAAGCAAATATGTGCGTTCTTCGTTGTCAGCTTCGAGTCTGACCCATGCCCGGATGTCCCGTCAATTCGGTGGCCAGTAGTTCAGGTTAGTCGGGTCTGTCTGTTCGTCAGGGAACAGTCTGTCTTCGAGACATTGTTTAGTTCCGAGCAGGACCCAAAATCACAGCAATTAGCGCGGAATTAACGACCGATTTCAACATG **************************************************************************************************************************************************.....((((((.....((((((..(((((...))))))))))).....))))))*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V120 male body |
M043 female body |
V058 head |
M056 embryo |
V052 head |
M026 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................CTGTCTGTTCGTCAGGGAACA.................................................................................... | 21 | 0 | 1 | 4.00 | 4 | 2 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................CGTCAGGGAACAGTCTGTCTT........................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GACATTGTTTAGTTCCGAGCA................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGTTAGTCGGGTCTGTCTGTTC............................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTCAGGGAACAGTCTGTCTTCGAGAC..................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AGTTCAGGTTAGTCGGGT......................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACCCAAAATCACAGCAATTAGCGCGGA...................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTCTTCGAGACATTGTTTAGTTCCGAGC.................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................AAAATCACAGCAATTAGCGCGGA...................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ATCACAGCAATTAGCGCGGAAT.................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................ATTGTTTAGTTCCGAGCAGG................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................GTCAGGGAACAGTCTGTCTT........................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GAACAGTCTGTCTTCGAGACATTGT................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTGTTTAGTTCCGAGCAGGA................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGTCGGGTCTGTCTGTTCGTC............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTCAGGGAACAGTCTGTCTTCGA........................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................AATTAGCGCGGAATTAACGACCG........... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................GTCTGTCTGTTCGTCAGGGAACAGTCTGT.............................................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTCGTCAGGGAACAGTCTGT.............................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................TCAGGGAACAGTCTGTCTTC.......................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CGGGTCTGTCTGTTCGTCAGGGAACA.................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................AAATCACAGCAATTAGCGCGG....................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................CAGCAATTAGCGCGGAATTAACGACC............ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTCGTCAGGGAACAGTCTGTCTTCGAGA...................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................GTTCGTCAGGGAACAGTCTGTCTTCGAGA...................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTCAGGGAACAGTCTGTC............................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................TCAGGTTAGTCGGGTCTGTCTGTT................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GACCCAAAATCACAGCAATTAGCGCGGAA..................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ATCACAGCAATTAGCGCGG....................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................CATTGTTTAGTTCCGAGCAG.................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GTAGTTCAGGTTAGTCGGGTCT....................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GTTCAGGTTAGTCGGGTCT....................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTCAGGGAACAGTCTGTCTTCGAGA...................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................ATCACAGCGATTAGCGCGGA...................... | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TCTTCGAAACTTTGTTTAGT........................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................AGTCTGACACCTGCCCGG.................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CTTCGAAACTTTGTTTAGTT.......................................................... | 20 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GGGTTTGACTGTTCGTCA........................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................CCCGGGTGTCCCCTCAA........................................................................................................................................ | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................GTAATTAGCGGGGAATTACC................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GTCTGACGCCTGCCCGGT................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
|
GACCGAGCTGTGACCAAAACAGCGGAATAGTCGATAGTCGATTACTAATGTTTCGTTTATACACGCAAGAAGCAACAGTCGAAGCTCAGACTGGGTACGGGCCTACAGGGCAGTTAAGCCACCGGTCATCAAGTCCAATCAGCCCAGACAGACAAGCAGTCCCTTGTCAGACAGAAGCTCTGTAACAAATCAAGGCTCGTCCTGGGTTTTAGTGTCGTTAATCGCGCCTTAATTGCTGGCTAAAGTTGTAC
***************************************************.....((((((.....((((((..(((((...))))))))))).....))))))************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V058 head |
M056 embryo |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................TAGTCCTGGGTTTTACTGGCGT................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 |
| .......................GGAATAGTCAATAGGCGTTT................................................................................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 |
| ............................................................................................................................................................................AGCAGCTTTGAAACAAATCA........................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droYak3 | 2R:20473576-20473826 - | dya_634 | CTGGCTCGA--CACTGG---TTTTGTCG-CCTTATCAGCTATCAGCTAATGATTACAAAGCAAATATGTGCGTTCTTC----------------------GTTGTCAGCTTC--GAGTCTGACCCATGCCCGGATGT-------------CCCGTCAAT------TCGGTGGCCAG-TAGTTCAGGTTAGTCGGGTCTGTCTG-----T-TCGTCAGGG-----AA-----CAGTCT-----GTCTTCGAGA--------------------------------------CATTGTTTAGTTCCGAGCA---GGA-----CCCAAA-----ATCA---------------------------------------CAGCAATTAGCGCGG-AATT------------AACGACCGATTT--------------------------------------------------------------------CAACATG |
| droEre2 | scaffold_4845:21876673-21876914 - | CTGGCTCGA--CACTGG---TATTGTCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGTTCTTC----------------------GTTGTCAGCTTC--GAGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TAGCTCAGGTTAGTCGGGTCTGTCTG-----T-TCGTCGGAG-----AA-----CGG-CT-----GTCT-CGAGA--------------------------------------CATTTCTTAGTTCCGATCA---GGA-----C-----CAAAAATCG---------------------------------------CAGCAATCAGCGCGC-AATT------------AACCCCCGATTT--------------------------------------------------------------------CAACATG | |
| droSec2 | scaffold_23:375324-375540 - | CTGGCTCGA--CACTGG---TTTTGTCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGTTCTTC----------------------GTTGTCAGCCCC--GAGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TAGTTCAGGTTAGTCGGCTCTGTCAG-----T-TCGTCAGAG-----AA-----CGG-GT-----GTCTTCGAGA--------------------------------------AGTTTTTCAGTTCCAATCA-------------------------------------------------------------------------------------------GGAATTAACACCCGAGTT--------------------------------------------------------------------CAACATG | |
| droSim2 | 2r:20981516-20981732 - | CTGGCTCGA--CACTGG---TTTTGTCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGTTCCCC----------------------GTTGTCAGCCCC--GAGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TAGTTCAGGTTAGTCGGCTCTGTCAG-----T-TCGTCAGAG-----AA-----CGG-CT-----GTCTTCGAGA--------------------------------------AGTTTTTCAATTCCAATCA-------------------------------------------------------------------------------------------GGAATTAACACCCGAGTT--------------------------------------------------------------------CAACATG | |
| dm3 | chr2R:20501796-20502012 - | CTGGCTCGA--CACTGG---TTTTGTCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGTTCTTC----------------------GCTGTCAGCCCC--GAGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TAGTTCAGGTTAGTCGGCTCTGTCTG-----T-TCGTCAGAG-----AA-----CTG-CT-----GTCTTCGAGA--------------------------------------CATTTTTCAGTTCCAATCA-------------------------------------------------------------------------------------------GGAATTAACACCCGAGTT--------------------------------------------------------------------CAACATG | |
| droEug1 | scf7180000409464:869843-870068 - | CTGGCTCGA--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTCTTG----------------------GTTTTCGGCCTG--GTGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTAGCCAG-TGGTTCAGGTTAGTCGGGTTCGTCGG-----T-TTGCCAGAG-----AA-----CGG-CT-----GTCTTCGAAGG-------------------------------------AGTTTCAA--------------------------AA-----ATCT---------------------------------------AATAAATCAGCGCGA-AGTT------------AATCCCCAATTT--------------------------------------------------------------------GAACATG | |
| droBia1 | scf7180000301754:3400253-3400514 + | CTGGCTCGA--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTCTTGGGTTTTGGTTTTTGCTATTT-GGTTTCTGGCCGGGTGTGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TGGTTCA-----GTCGGGTTTGTCGG-----T-TCGTCAGGG-----AA-----CGG-CG-----GTCTTCGAGAG-------------------------------------AGTTTCTAAAATTCGTTAG---GCA-----T-----CGCAAGTAG---------------------------------------CCCTAATCAGCGCGG-AGTT------------AAGCCCCAATTT--------------------------------------------------------------------AAACATG | |
| droTak1 | scf7180000414175:10517-10741 + | CTGGCTCGA--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTCTTC---------------------GGTTTTTGGTCTGGTATGCCTGACCCATGCCCGGATGT-------------CCTATCAATT-----TCGGTGGCCAG-TGGTTCA-----GTCGGCTACCTCGG-----T-TCGTCAGGG-----AA-----AGA---------------------------------------------------------GTTTCT------TGTTTAGAC-AACG-----AAAA-----ATCA---------------------------------------CATAAATCAGCGCGT-AATT------------AATCCCCAATTT--------------------------------------------------------------------AAACATG | |
| droEle1 | scf7180000491214:4478761-4478983 + | CTGGCTCGA--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAACGATTACAAAGCAAATATGTGTGTTCTTGGTT-TTGGTTT--------G-GGTTTTTGGCCTG--CTGTCTGACCCATGCCCGGATGT-------------CCTATCAATT-----TCGGTGGCCAG-TAGTGCAGGTTAGTCGGGTTCGTCAG-----T-TTGTCGGAG-----AA-----CGG-TT-----GTCTTCAA--------------------------C----------------TTCAA------------------------CAAA-----ATCG---------------------------------------TATCGATCAGCGCGT-AG-------------------------T--------------------------------------------------------------------AAAAATG | |
| droRho1 | scf7180000776849:203099-203351 - | CTGGCTCGA--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTTTTGGTTTT---------------TGGTTTTTGGCCTG--GTGTCTGACCCATGCCCGGATGT-------------CCTATCAAT------TCGGTGGCCAG-TAGTTCATGTTAGTCGGGTTCGTCAG-----T-TTGTCAGAG-----AA-----CGG-TG-----GTCTCCGAAGGAA------------------------------AGT--GGAATCTA-AATTCGATCA---ATC-----C-----CGAA-AGCA---------------------------------------CATTAATTAGTGCGGTA--T-----AGTAGTTA-------ACTT--------------------------------------------------------------------AACCATG | |
| droFic1 | scf7180000453342:356337-356582 - | CTGGCTCGA--CACTGG---TTTTGTCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTCTTG----------------------GTTTTCGGCCTG--CCGTCTGACCCATGCCCGGATGT-------------CCCGTCAAT------TCGGTGGCCAG-TAGCTCAGGTTAGTCGGGTTGGTCGG-----T-TCGATAGAG-----AA-----CGG-TT-----GTCTGAAAGG--------------------------------------GGTTTCTA-AATTTGTTCA---GTA-----C-----CGAAAATCG---------------------------------------CAAAAGTCGGCGCGC-AATTAATCC-GGAAATA-------ATTT--------------------------------------------------------------------AAACATG | |
| droKik1 | scf7180000302470:1905844-1906085 + | CTGGCTCGG--CACTGG---TTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTC----------------------------GTTAT-----TG--GTGTCTGACCCACGCCCCGATGA-----------GGGCCATCAAT------GCGGTGGCCAG-TAGCTCGGGTTAGTTGGGTTCGACGG-----T-TCGCCAAAG---------------------------TCGAAGCAAGTTTCACCTTTTTGTTT----TT--------------------------CTTCAGAC---CGAAACTCTGA-----ATCATAATTTAGAGTGGAAAAATCCCCTAACTAAAGTCC----------------------------------------------CT--------------------------------------------------------------------TAACATG | |
| droAna3 | scaffold_13266:7092520-7092768 - | CTGGCTAGG--C-------------CCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGTTTTTT----T---------------TCGATTTTATTGTA--GTATCTGTCTCATGTCCGCCCGG-------------GCCGTCGAT------TCGGTAGCCAG-TACTTCATGTTAGTTGGGTTCGTCAG-TCGGT------ACTGTCGAGCAGGAGC------------------------GTTCCTCCA------------TTCCTCATAAATCGAGCGTC------------G---AAA-----T-----CGAAAATCG-------------CAA-------------AAGCCCCGTATATTAATCAGAACAA-A--T-----AGTCATTA----------A--------------------------------------------------------------------AAAAATG | |
| droBip1 | scf7180000396759:91024-91270 + | CTGGCTAGG--C-------------TCG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGTGTTTTTT--------------------TCGATTTTATTATG--GTATCTGTCCCGTGTCTGCCTGG-------------GCCGTCGAT------TCGGTAGCCAG-TACTTCTTGTTAGTTTGGTTCGTCAG-TCGGT------ACTGTCGAGCAGGAGT------------------------GTTCCTCCA------------TTCCTCAAAAATCGAGTATC------------G---AAA-----T-----CGAAAACCG-------------CTA-------------AAGCCCC-CACATTAATCAGAACAA-A--T-----AGTCATTA----------A--------------------------------------------------------------------AAATATG | |
| dp5 | 3:17197983-17198239 + | CTGGCTCGGGTTGCTGC---CTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGGGTGTGCGG-CTGGTGT--------GCGGCGGGTGTGCGC--AGGTCTGACCCATGCCCGGTTGG------------GGCCATCAAG------TTGG--GCCAG--------------TCTAGTTAGTCAG-----T-TCGATCGAG-----CT-----CCG-CCGCGTCGCCTTTGAAGCAATTGATACACTTCGATTCGTGTTA-CGC--------------------------A---AA-----------------ATAATAAATA---------------------------------TATAGATAGACAAGA-AATT------------A-T-ATCGAGTG--------------------------------------------------------------------GAATATG | |
| droPer2 | scaffold_31:591946-592191 - | CTGGCTCGGGTTGCTAC---CTTTGTCA-CCTTAT-------CAGCTAATGATTACAAAGCAAATATGTGCGGGTGTG--------------------CGGCGGGTGTGCGC--AGGTCTGACCCATGCCCGGTTGG------------GGCCATCAAT------TTGG--GCCAG--------------TCTAGTTAGTCAG-----T-TCGATCGAG-----CT-----CCG-CCGCGTCGCCTTTGAAGCAATTGATACACTTCGATTAGTGTTA-CGC--------------------------A---AA-----------------ATAATAGATA---------------------------------GATAGATAGACAAGA-AATT------------A-T-ATCGAGTG--------------------------------------------------------------------GAATATG | |
| droWil2 | scf2_1100000004513:4533937-4534175 + | CTTGTAAGC--TACTGAAATCCTTCATG-CCTTAT-------CAGCTAATGATTACAAAGCAAATATCTCTCT---------------------------GTCTGTATAGGT--GAACCTGACCCATGGGGAGGTGTTTGGTCG--GGCAATCATCAAT------TTGTCGGTCAGGTTTTTTTCTTTAGTTTCGATTT-TGATTCACTAACGAGCAAA-----CG-----CCC-ACGCAACGACACGGA---AATACATATATTTCGG-------------------------------------------------------------------------------------------------------------------TTCAA-G---------------------------------------------------------------------TTTTCAGTCTTAAAAAAAA-------AAGAATG | |
| droVir3 | scaffold_12875:964656-964892 - | CAGGCTAGG--CACTGG---TTTTGTCAACTTTAT-------CAGCTAATGATTACAAAGCAAATATGTGAGC--------------------------------------C--GTGCCTGACCCATGATGA-CTGG----TCGCCGGGGCCCATCAATTTATTTTCGGCGAC-TT-CAGTTCAATTCAGTTCGAACGGACAC-----T-TTGAGTGAA-----A------CGA---------------------------------------------------------GA--------------------------------------------------------------------------------------------------------------------------CTTGTGCAAAACTATTTTTATAACCGAAATCAAGTCCGATACAAATGG-CGACCGCAACAATAACAGCATCAACCGCT | |
| droMoj3 | scaffold_6496:4002971-4003125 - | TTTTTTC---------T---TTTTGTCAACCTTAT-------CAGCTAATGATTACAAAGCAAACATGTGAA------------------------------------CCAT--GAGCCTGACCCATGGATGAGTGG----TCG--GTCGCCCATCAAT------TTGGGGGGCTT-CAGTTCAATTCGAGCGGACACAACG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTA-----------------------GAAACAATATTT--------------------------------------------------------------------CTGAATG | |
| droGri2 | scaffold_15245:10002642-10002717 + | CCGACTTGG--CACTGA---TTTTGTCAACTTTAT-------CAGCTAATGATTACAAAGCAAATATGTGAGC--------------------------------------C--GTGCCTGACCCATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 10:55 PM