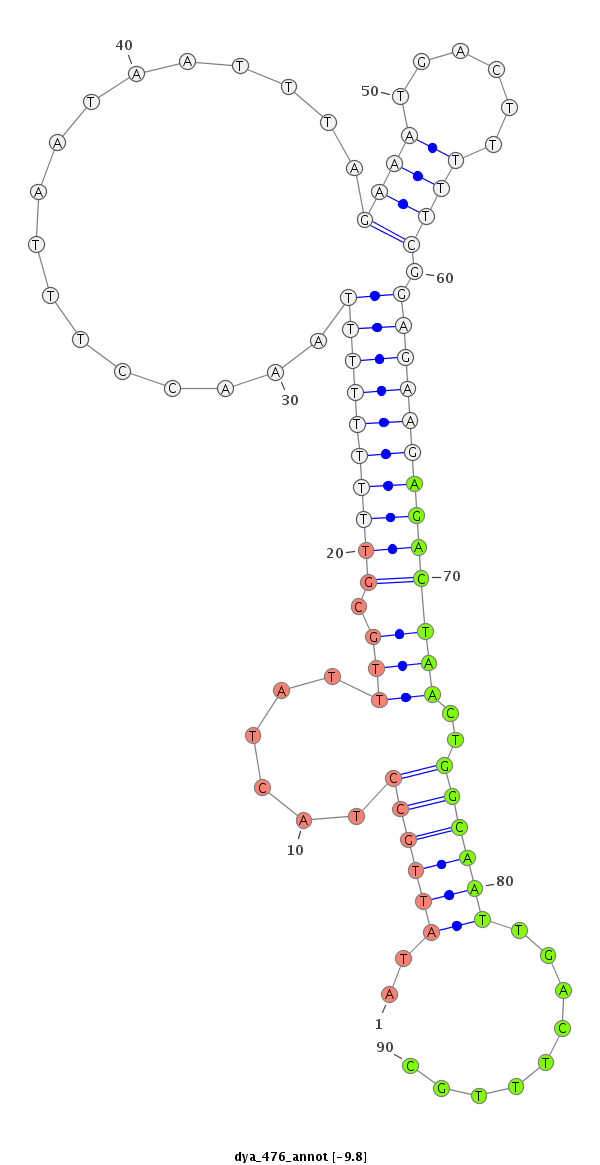
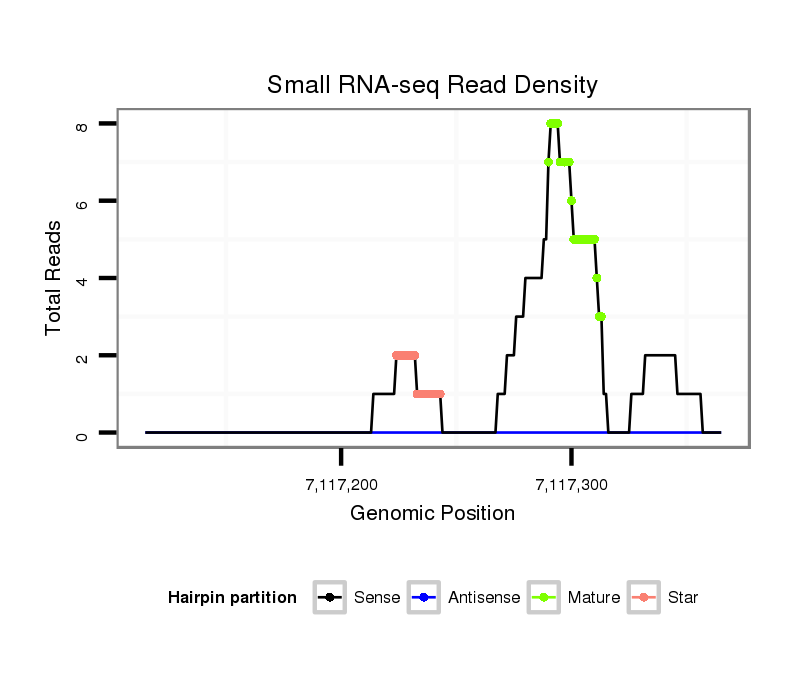
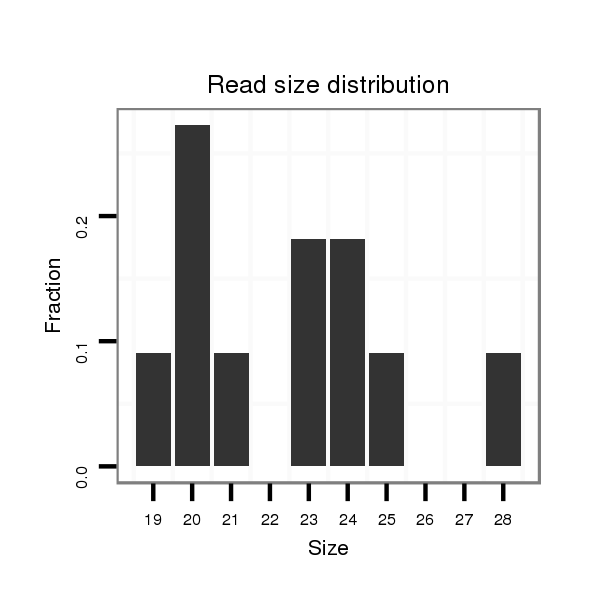
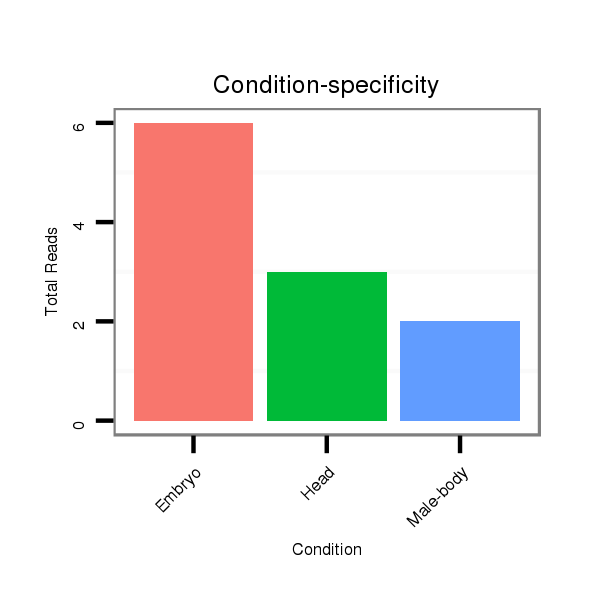
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACCA..... |
18 |
1 |
2 |
242.50 |
485 |
168 |
124 |
79 |
99 |
10 |
0 |
5 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGTCACCA..... |
19 |
1 |
1 |
86.00 |
86 |
38 |
22 |
8 |
13 |
2 |
3 |
0 |
0 |
| ..................................................................................................................................................................................................................................GAATCCCACCGCTGTCACCA..... |
20 |
2 |
2 |
42.00 |
84 |
25 |
7 |
40 |
4 |
3 |
5 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCA..... |
17 |
1 |
4 |
5.25 |
21 |
0 |
21 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................TATCCCACCGCTGTCACCA..... |
19 |
2 |
4 |
3.00 |
12 |
4 |
5 |
2 |
0 |
0 |
1 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGCCACCA..... |
19 |
2 |
13 |
2.85 |
37 |
29 |
1 |
4 |
0 |
0 |
1 |
2 |
0 |
| ................................................................................................................................................................................................................................TTGAATCCCACCGCTGTCACCA..... |
22 |
3 |
2 |
2.50 |
5 |
1 |
0 |
3 |
0 |
0 |
1 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGTCACC...... |
18 |
1 |
2 |
2.00 |
4 |
1 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................AGACTAACTGGCAATTGACTTTGC.................................................... |
24 |
0 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................TTAATCCCACCGCTGTCACCA..... |
21 |
2 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................GTATCCCACCGCTGTCACCA..... |
20 |
3 |
12 |
1.67 |
20 |
5 |
0 |
14 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACCC..... |
18 |
2 |
3 |
1.33 |
4 |
0 |
3 |
0 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................................................................................................................GACTAACTGGCAATTGACTTT...................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................TCGATCCCACCGCTGTCACCA..... |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................ATATTGCCTACTATTTGCGT.......................................................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .........................................................................................................................................................................................................................CTCGTCGGTCAATCCCACCGCCGTC......... |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...................................................................................................................................................................................................................GCCCGGCTCGTCGGTCAATC.................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................GACTAACTGGCAATTGACTT....................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................TCCAATCCCACCGCTGTCACCA..... |
22 |
3 |
2 |
1.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................GTACTGTTAGATATTGCCT..................................................................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..............................................................................................................................................................................................................................CGATCGATCCCACCGCGGTCACCA..... |
24 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CTTTTTCGGAGAAGAGACTAACTGG................................................................. |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCAG.... |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................AGAGACTAACTGGCAATTGACTTTGCAG.................................................. |
28 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................AGCCCACCGCCGTCACCA..... |
18 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................TTCGGAGAAGAGACTAACTG.................................................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .........................................................................................................................................................AGAAATGACTTTTTCGGAGAAGA........................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ................................................................................................................................................................................................................................GGGAATCCCACCGCTGTCACCA..... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCAGT... |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................ATGACTTTTTCGGAGAAGAGACT....................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AGTCCCACCGCTGTCACCA..... |
19 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCATT... |
19 |
2 |
8 |
0.88 |
7 |
5 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCAA.... |
18 |
2 |
11 |
0.82 |
9 |
7 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................GATCCCACCGCTGTCACCA..... |
19 |
2 |
4 |
0.75 |
3 |
0 |
0 |
2 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................TCGAATCCCACCGCCGTCACCA..... |
22 |
3 |
3 |
0.67 |
2 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCCCCA..... |
18 |
2 |
9 |
0.67 |
6 |
0 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................CGAATCCCACCGCCGTCACC...... |
20 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCCCTGTCACCA..... |
19 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGTCCCCA..... |
19 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCTACCGCTGTCACCA..... |
19 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................CAATCCCACCGCTGCCACCA..... |
20 |
2 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCTCTGTCACCA..... |
18 |
2 |
7 |
0.43 |
3 |
1 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACAGCTGTCACCA..... |
19 |
2 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................TAAAATCCCACCGCCGTCACCA..... |
22 |
3 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGTCGCCA..... |
19 |
2 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................GCGAATCCCACCGCTGTCACCA..... |
22 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACCATT... |
20 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACCT..... |
18 |
2 |
10 |
0.30 |
3 |
0 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................GGATCCCACCGCCGTCACCA..... |
20 |
2 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................GTAATCCCACCGCTGTCACCA..... |
21 |
3 |
4 |
0.25 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCCGCCACCA..... |
18 |
1 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ....................................................................................GAAACAACCCAGACCGTC..................................................................................................................................................... |
18 |
3 |
20 |
0.20 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACGA..... |
18 |
2 |
5 |
0.20 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCACCAA.... |
19 |
2 |
5 |
0.20 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................GTCCCACCGCTGTCACCA..... |
18 |
2 |
11 |
0.18 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................CAAATCCCACCGCTGTCACCA..... |
21 |
3 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCACTCACCA..... |
18 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................GATCCCACCGCTGTCACC...... |
18 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCACTGTCACCA..... |
18 |
2 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATACCACCGCAGTCACCA..... |
18 |
2 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCAACA..... |
18 |
2 |
8 |
0.13 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCACCGCTGTCGCCA..... |
18 |
2 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCTCACCGCTGTCACCA..... |
18 |
2 |
9 |
0.11 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATCCCAACGCTGTCACCA..... |
18 |
2 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................GAGTCCCACCGCTGTCACCA..... |
20 |
3 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCGCTGGCACC...... |
18 |
2 |
13 |
0.08 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TCCCACCGCTGTCACCAC.... |
18 |
2 |
15 |
0.07 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................AATCCCACCTCTGACACCA..... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................................................CCACCGCTGTCACCA..... |
15 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ................................................................GGATTGATCCAGACATGT......................................................................................................................................................................... |
18 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................................................CACCGCCGTCGCTAGTAC. |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |